Text mining of full-text journal articles combined with gene expression analysis reveals a relationship between sphingosine-1-phosphate and invasiveness of a glioblastoma cell line
- PMID: 16901352
- PMCID: PMC1557675
- DOI: 10.1186/1471-2105-7-373
Text mining of full-text journal articles combined with gene expression analysis reveals a relationship between sphingosine-1-phosphate and invasiveness of a glioblastoma cell line
Abstract
Background: Sphingosine 1-phosphate (S1P), a lysophospholipid, is involved in various cellular processes such as migration, proliferation, and survival. To date, the impact of S1P on human glioblastoma is not fully understood. Particularly, the concerted role played by matrix metalloproteinases (MMP) and S1P in aggressive tumor behavior and angiogenesis remains to be elucidated.
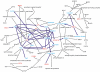
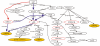
Results: To gain new insights in the effect of S1P on angiogenesis and invasion of this type of malignant tumor, we used microarrays to investigate the gene expression in glioblastoma as a response to S1P administration in vitro. We compared the expression profiles for the same cell lines under the influence of epidermal growth factor (EGF), an important growth factor. We found a set of 72 genes that are significantly differentially expressed as a unique response to S1P. Based on the result of mining full-text articles from 20 scientific journals in the field of cancer research published over a period of five years, we inferred gene-gene interaction networks for these 72 differentially expressed genes. Among the generated networks, we identified a particularly interesting one. It describes a cascading event, triggered by S1P, leading to the transactivation of MMP-9 via neuregulin-1 (NRG-1), vascular endothelial growth factor (VEGF), and the urokinase-type plasminogen activator (uPA). This interaction network has the potential to shed new light on our understanding of the role played by MMP-9 in invasive glioblastomas.
Conclusion: Automated extraction of information from biological literature promises to play an increasingly important role in biological knowledge discovery. This is particularly true for high-throughput approaches, such as microarrays, and for combining and integrating data from different sources. Text mining may hold the key to unraveling previously unknown relationships between biological entities and could develop into an indispensable instrument in the process of formulating novel and potentially promising hypotheses.
Figures




Similar articles
-
Sphingosine-1-phosphate regulates glioblastoma cell invasiveness through the urokinase plasminogen activator system and CCN1/Cyr61.Mol Cancer Res. 2009 Jan;7(1):23-32. doi: 10.1158/1541-7786.MCR-08-0061. Mol Cancer Res. 2009. PMID: 19147534 Free PMC article.
-
A metabolic shift favoring sphingosine 1-phosphate at the expense of ceramide controls glioblastoma angiogenesis.J Biol Chem. 2013 Dec 27;288(52):37355-64. doi: 10.1074/jbc.M113.494740. Epub 2013 Nov 21. J Biol Chem. 2013. PMID: 24265321 Free PMC article. Clinical Trial.
-
Modulation of invasive properties of CD133+ glioblastoma stem cells: a role for MT1-MMP in bioactive lysophospholipid signaling.Mol Carcinog. 2009 Oct;48(10):910-9. doi: 10.1002/mc.20541. Mol Carcinog. 2009. PMID: 19326372
-
Sphingolipid Metabolism in Glioblastoma and Metastatic Brain Tumors: A Review of Sphingomyelinases and Sphingosine-1-Phosphate.Biomolecules. 2020 Sep 23;10(10):1357. doi: 10.3390/biom10101357. Biomolecules. 2020. PMID: 32977496 Free PMC article. Review.
-
Sphingosin 1-phosphate contributes in tumor progression.J Cancer Res Ther. 2013 Oct-Dec;9(4):556-63. doi: 10.4103/0973-1482.126446. J Cancer Res Ther. 2013. PMID: 24518696 Review.
Cited by
-
The Text-mining based PubChem Bioassay neighboring analysis.BMC Bioinformatics. 2010 Nov 8;11:549. doi: 10.1186/1471-2105-11-549. BMC Bioinformatics. 2010. PMID: 21059237 Free PMC article.
-
Seeking a new biology through text mining.Cell. 2008 Jul 11;134(1):9-13. doi: 10.1016/j.cell.2008.06.029. Cell. 2008. PMID: 18614002 Free PMC article.
-
Networking of differentially expressed genes in human cancer cells resistant to methotrexate.Genome Med. 2009 Sep 4;1(9):83. doi: 10.1186/gm83. Genome Med. 2009. PMID: 19732436 Free PMC article.
-
Sphingosine-1-Phosphate in the Tumor Microenvironment: A Signaling Hub Regulating Cancer Hallmarks.Cells. 2020 Feb 1;9(2):337. doi: 10.3390/cells9020337. Cells. 2020. PMID: 32024090 Free PMC article. Review.
-
Protein kinase D2 regulates migration and invasion of U87MG glioblastoma cells in vitro.Exp Cell Res. 2013 Aug 1;319(13):2037-2048. doi: 10.1016/j.yexcr.2013.03.029. Epub 2013 Apr 4. Exp Cell Res. 2013. PMID: 23562655 Free PMC article.
References
-
- Malek RL, Toman RE, Edsall LC, Wong S, Chiu J, Letterle CA, Van Brocklyn JR, Milstien S, Spiegel S, Lee NH. Nrg-1 belongs to the endothelial differentiation gene family of G protein-coupled sphingosine-1-phosphate receptors. J Biol Chem. 2001;276:5692–5699. doi: 10.1074/jbc.M003964200. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

