A high-density consensus map of barley linking DArT markers to SSR, RFLP and STS loci and agricultural traits
- PMID: 16904008
- PMCID: PMC1564146
- DOI: 10.1186/1471-2164-7-206
A high-density consensus map of barley linking DArT markers to SSR, RFLP and STS loci and agricultural traits
Abstract
Background: Molecular marker technologies are undergoing a transition from largely serial assays measuring DNA fragment sizes to hybridization-based technologies with high multiplexing levels. Diversity Arrays Technology (DArT) is a hybridization-based technology that is increasingly being adopted by barley researchers. There is a need to integrate the information generated by DArT with previous data produced with gel-based marker technologies. The goal of this study was to build a high-density consensus linkage map from the combined datasets of ten populations, most of which were simultaneously typed with DArT and Simple Sequence Repeat (SSR), Restriction Enzyme Fragment Polymorphism (RFLP) and/or Sequence Tagged Site (STS) markers.
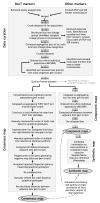
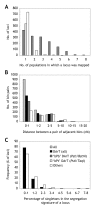
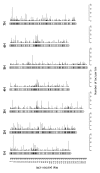
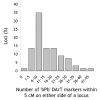
Results: The consensus map, built using a combination of JoinMap 3.0 software and several purpose-built perl scripts, comprised 2,935 loci (2,085 DArT, 850 other loci) and spanned 1,161 cM. It contained a total of 1,629 'bins' (unique loci), with an average inter-bin distance of 0.7 +/- 1.0 cM (median = 0.3 cM). More than 98% of the map could be covered with a single DArT assay. The arrangement of loci was very similar to, and almost as optimal as, the arrangement of loci in component maps built for individual populations. The locus order of a synthetic map derived from merging the component maps without considering the segregation data was only slightly inferior. The distribution of loci along chromosomes indicated centromeric suppression of recombination in all chromosomes except 5H. DArT markers appeared to have a moderate tendency toward hypomethylated, gene-rich regions in distal chromosome areas. On the average, 14 +/- 9 DArT loci were identified within 5 cM on either side of SSR, RFLP or STS loci previously identified as linked to agricultural traits.
Conclusion: Our barley consensus map provides a framework for transferring genetic information between different marker systems and for deploying DArT markers in molecular breeding schemes. The study also highlights the need for improved software for building consensus maps from high-density segregation data of multiple populations.
Figures



References
-
- Zohary D, Hopf M. Domestication of plants in the old world. Oxford, United Kingdom: Clarendon Press; 1988.
-
- Vivar HE, McNab A. Proceedings of an International Symposium: 13–14 March 2000. Ciudad Obregon, Mexico. CIMMYT; 2003. Breeding barley in the new millemium.
-
- Langridge P, Barr AR. Preface. Austr J Agric Res. 2003;54:i–iv. doi: 10.1071/ARv54n12_PR. - DOI
-
- Hayes PM, Castro A, Marquez-Cedillo L, Corey A, Henson C, Jones B, Kling J, Mather D, Matus I, Rossi C, Sato K. A summary of published barley QTL reports http://www.barleyworld.org/northamericanbarley/qtlsummary.php
-
- Koebner RMD, Powell W, Donini P. Contributions of DNA molecular marker technologies to the genetics and breeding of what and barley. Plant Breed Rev. 2001;21:181–220.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

