Dynamically driven protein allostery
- PMID: 16906160
- PMCID: PMC2757644
- DOI: 10.1038/nsmb1132
Dynamically driven protein allostery
Abstract
Allosteric interactions are typically considered to proceed through a series of discrete changes in bonding interactions that alter the protein conformation. Here we show that allostery can be mediated exclusively by transmitted changes in protein motions. We have characterized the negatively cooperative binding of cAMP to the dimeric catabolite activator protein (CAP) at discrete conformational states. Binding of the first cAMP to one subunit of a CAP dimer has no effect on the conformation of the other subunit. The dynamics of the system, however, are modulated in a distinct way by the sequential ligand binding process, with the first cAMP partially enhancing and the second cAMP completely quenching protein motions. As a result, the second cAMP binding incurs a pronounced conformational entropic penalty that is entirely responsible for the observed cooperativity. The results provide strong support for the existence of purely dynamics-driven allostery.
Conflict of interest statement
Figures

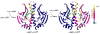



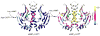
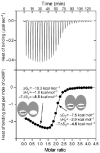

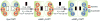
References
-
- Hardy JA, Wells JA. Searching for new allosteric sites in enzymes. Curr Opin Struct Biol. 2004;14:706–715. - PubMed
-
- Swain JF, Gierasch LM. The changing landscape of protein allostery. Curr Opin Struct Biol. 2006;16:102–108. - PubMed
-
- Suel GM, Lockless SW, Wall MA, Ranganathan R. Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat Struct Biol. 2003;10:59–69. - PubMed
-
- Koshland DE., Jr Conformational changes: how small is big enough. Nat Med. 1998;4:1112–1114. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

