Critical points of DNA quantification by real-time PCR--effects of DNA extraction method and sample matrix on quantification of genetically modified organisms
- PMID: 16907967
- PMCID: PMC1569826
- DOI: 10.1186/1472-6750-6-37
Critical points of DNA quantification by real-time PCR--effects of DNA extraction method and sample matrix on quantification of genetically modified organisms
Abstract
Background: Real-time PCR is the technique of choice for nucleic acid quantification. In the field of detection of genetically modified organisms (GMOs) quantification of biotech products may be required to fulfil legislative requirements. However, successful quantification depends crucially on the quality of the sample DNA analyzed. Methods for GMO detection are generally validated on certified reference materials that are in the form of powdered grain material, while detection in routine laboratories must be performed on a wide variety of sample matrixes. Due to food processing, the DNA in sample matrixes can be present in low amounts and also degraded. In addition, molecules of plant origin or from other sources that affect PCR amplification of samples will influence the reliability of the quantification. Further, the wide variety of sample matrixes presents a challenge for detection laboratories. The extraction method must ensure high yield and quality of the DNA obtained and must be carefully selected, since even components of DNA extraction solutions can influence PCR reactions. GMO quantification is based on a standard curve, therefore similarity of PCR efficiency for the sample and standard reference material is a prerequisite for exact quantification. Little information on the performance of real-time PCR on samples of different matrixes is available.
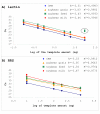
Results: Five commonly used DNA extraction techniques were compared and their suitability for quantitative analysis was assessed. The effect of sample matrix on nucleic acid quantification was assessed by comparing 4 maize and 4 soybean matrixes. In addition 205 maize and soybean samples from routine analysis were analyzed for PCR efficiency to assess variability of PCR performance within each sample matrix. Together with the amount of DNA needed for reliable quantification, PCR efficiency is the crucial parameter determining the reliability of quantitative results, therefore it was chosen as the primary criterion by which to evaluate the quality and performance on different matrixes and extraction techniques. The effect of PCR efficiency on the resulting GMO content is demonstrated.
Conclusion: The crucial influence of extraction technique and sample matrix properties on the results of GMO quantification is demonstrated. Appropriate extraction techniques for each matrix need to be determined to achieve accurate DNA quantification. Nevertheless, as it is shown that in the area of food and feed testing matrix with certain specificities is impossible to define strict quality controls need to be introduced to monitor PCR. The results of our study are also applicable to other fields of quantitative testing by real-time PCR.
Figures




References
-
- James C. Highlights of ISAAA briefs no. 34-2005 Global status of commercialized biotech/GM crops. ISAAA. 2005.
-
- Holst-Jensen A, Ronning SB, Lovseth A, Berdal KG. PCR technology for screening and quantification of genetically modified organisms (GMOs) Anal Bioanal Chem. 2003;375:985–993. - PubMed
-
- Lipp M, Shillito R, Giroux R, Spiegelhalter F, Charlton S, Pinero D, Song P. Polymerase chain reaction technology as analytical tool in agricultural biotechnology. J AOAC Int. 2005;88:136–155. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

