The origin of introns and their role in eukaryogenesis: a compromise solution to the introns-early versus introns-late debate?
- PMID: 16907971
- PMCID: PMC1570339
- DOI: 10.1186/1745-6150-1-22
The origin of introns and their role in eukaryogenesis: a compromise solution to the introns-early versus introns-late debate?
Abstract
Background: Ever since the discovery of 'genes in pieces' and mRNA splicing in eukaryotes, origin and evolution of spliceosomal introns have been considered within the conceptual framework of the 'introns early' versus 'introns late' debate. The 'introns early' hypothesis, which is closely linked to the so-called exon theory of gene evolution, posits that protein-coding genes were interrupted by numerous introns even at the earliest stages of life's evolution and that introns played a major role in the origin of proteins by facilitating recombination of sequences coding for small protein/peptide modules. Under this scenario, the absence of spliceosomal introns in prokaryotes is considered to be a result of "genome streamlining". The 'introns late' hypothesis counters that spliceosomal introns emerged only in eukaryotes, and moreover, have been inserted into protein-coding genes continuously throughout the evolution of eukaryotes. Beyond the formal dilemma, the more substantial side of this debate has to do with possible roles of introns in the evolution of eukaryotes.
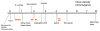
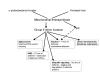
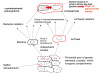
Results: I argue that several lines of evidence now suggest a coherent solution to the introns-early versus introns-late debate, and the emerging picture of intron evolution integrates aspects of both views although, formally, there seems to be no support for the original version of introns-early. Firstly, there is growing evidence that spliceosomal introns evolved from group II self-splicing introns which are present, usually, in small numbers, in many bacteria, and probably, moved into the evolving eukaryotic genome from the alpha-proteobacterial progenitor of the mitochondria. Secondly, the concept of a primordial pool of 'virus-like' genetic elements implies that self-splicing introns are among the most ancient genetic entities. Thirdly, reconstructions of the ancestral state of eukaryotic genes suggest that the last common ancestor of extant eukaryotes had an intron-rich genome. Thus, it appears that ancestors of spliceosomal introns, indeed, have existed since the earliest stages of life's evolution, in a formal agreement with the introns-early scenario. However, there is no evidence that these ancient introns ever became widespread before the emergence of eukaryotes, hence, the central tenet of introns-early, the role of introns in early evolution of proteins, has no support. However, the demonstration that numerous introns invaded eukaryotic genes at the outset of eukaryotic evolution and that subsequent intron gain has been limited in many eukaryotic lineages implicates introns as an ancestral feature of eukaryotic genomes and refutes radical versions of introns-late. Perhaps, most importantly, I argue that the intron invasion triggered other pivotal events of eukaryogenesis, including the emergence of the spliceosome, the nucleus, the linear chromosomes, the telomerase, and the ubiquitin signaling system. This concept of eukaryogenesis, in a sense, revives some tenets of the exon hypothesis, by assigning to introns crucial roles in eukaryotic evolutionary innovation.
Conclusion: The scenario of the origin and evolution of introns that is best compatible with the results of comparative genomics and theoretical considerations goes as follows: self-splicing introns since the earliest stages of life's evolution--numerous spliceosomal introns invading genes of the emerging eukaryote during eukaryogenesis--subsequent lineage-specific loss and gain of introns. The intron invasion, probably, spawned by the mitochondrial endosymbiont, might have critically contributed to the emergence of the principal features of the eukaryotic cell. This scenario combines aspects of the introns-early and introns-late views.
Reviewers: this article was reviewed by W. Ford Doolittle, James Darnell (nominated by W. Ford Doolittle), William Martin, and Anthony Poole.
Figures
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

