Cell organisation, sulphur metabolism and ion transport-related genes are differentially expressed in Paracoccidioides brasiliensis mycelium and yeast cells
- PMID: 16907987
- PMCID: PMC1578568
- DOI: 10.1186/1471-2164-7-208
Cell organisation, sulphur metabolism and ion transport-related genes are differentially expressed in Paracoccidioides brasiliensis mycelium and yeast cells
Abstract
Background: Mycelium-to-yeast transition in the human host is essential for pathogenicity by the fungus Paracoccidioides brasiliensis and both cell types are therefore critical to the establishment of paracoccidioidomycosis (PCM), a systemic mycosis endemic to Latin America. The infected population is of about 10 million individuals, 2% of whom will eventually develop the disease. Previously, transcriptome analysis of mycelium and yeast cells resulted in the assembly of 6,022 sequence groups. Gene expression analysis, using both in silico EST subtraction and cDNA microarray, revealed genes that were differential to yeast or mycelium, and we discussed those involved in sugar metabolism. To advance our understanding of molecular mechanisms of dimorphic transition, we performed an extended analysis of gene expression profiles using the methods mentioned above.
Results: In this work, continuous data mining revealed 66 new differentially expressed sequences that were MIPS(Munich Information Center for Protein Sequences)-categorised according to the cellular process in which they are presumably involved. Two well represented classes were chosen for further analysis: (i) control of cell organisation - cell wall, membrane and cytoskeleton, whose representatives were hex (encoding for a hexagonal peroxisome protein), bgl (encoding for a 1,3-beta-glucosidase) in mycelium cells; and ags (an alpha-1,3-glucan synthase), cda (a chitin deacetylase) and vrp (a verprolin) in yeast cells; (ii) ion metabolism and transport - two genes putatively implicated in ion transport were confirmed to be highly expressed in mycelium cells - isc and ktp, respectively an iron-sulphur cluster-like protein and a cation transporter; and a putative P-type cation pump (pct) in yeast. Also, several enzymes from the cysteine de novo biosynthesis pathway were shown to be up regulated in the yeast form, including ATP sulphurylase, APS kinase and also PAPS reductase.
Conclusion: Taken together, these data show that several genes involved in cell organisation and ion metabolism/transport are expressed differentially along dimorphic transition. Hyper expression in yeast of the enzymes of sulphur metabolism reinforced that this metabolic pathway could be important for this process. Understanding these changes by functional analysis of such genes may lead to a better understanding of the infective process, thus providing new targets and strategies to control PCM.
Figures

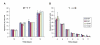

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

