Mitochondrial DNA-deletion mutations accumulate intracellularly to detrimental levels in aged human skeletal muscle fibers
- PMID: 16909385
- PMCID: PMC1559550
- DOI: 10.1086/507132
Mitochondrial DNA-deletion mutations accumulate intracellularly to detrimental levels in aged human skeletal muscle fibers
Abstract
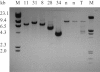
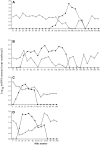
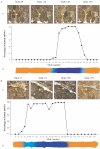
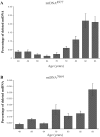
Skeletal muscle-mass loss with age has severe health consequences, yet the molecular basis of the loss remains obscure. Although mitochondrial DNA (mtDNA)-deletion mutations have been shown to accumulate with age, for these aberrant genomes to be physiologically relevant, they must accumulate to high levels intracellularly and be present in a significant number of cells. We examined mtDNA-deletion mutations in vastus lateralis (VL) muscle of human subjects aged 49-93 years, using both histologic and polymerase-chain-reaction (PCR) analyses, to determine the physiological and genomic integrity of mitochondria in aging human muscle. The number of VL muscle fibers exhibiting mitochondrial electron-transport-system (ETS) abnormalities increased from an estimated 6% at age 49 years to 31% at age 92 years. We analyzed the mitochondrial genotype of 48 single ETS-abnormal, cytochrome c oxidase-negative/succinate dehydrogenase-hyperreactive (COX-/SDH++) fibers from normal aging human subjects and identified mtDNA-deletion mutations in all abnormal fibers. Deletion mutations were clonal within a fiber and concomitant to the COX-/SDH++ region. Quantitative PCR analysis of wild-type and deletion-containing mtDNA genomes within ETS-abnormal regions of single fibers demonstrated that these deletion mutations accumulate to detrimental levels (>90% of the total mtDNA).
Figures


References
Web Resources
-
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for ND5 [accession number 2052366], Cytb [accession number AAB58955], COXI [accession number AAB58945], COXIII [accession number 2052365], and HVS-1 [accession number 1944628])
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CPEO) - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

