Meiotic recombination and spatial proximity in the etiology of the recurrent t(11;22)
- PMID: 16909390
- PMCID: PMC1559541
- DOI: 10.1086/507652
Meiotic recombination and spatial proximity in the etiology of the recurrent t(11;22)
Abstract
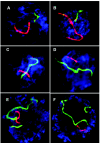
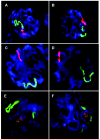
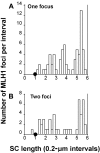
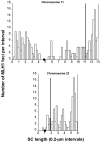
Although balanced translocations are among the most common human chromosomal aberrations, the constitutional t(11;22)(q23;q11) is the only known recurrent non-Robertsonian translocation. Evidence indicates that de novo formation of the t(11;22) occurs during meiosis. To test the hypothesis that spatial proximity of chromosomes 11 and 22 in meiotic prophase oocytes and spermatocytes plays a role in the rearrangement, the positions of the 11q23 and 22q11 translocation breakpoints were examined. Fluorescence in situ hybridization with use of DNA probes for these sites demonstrates that 11q23 is closer to 22q11 in meiosis than to a control at 6q26. Although chromosome 21p11, another control, often lies as close to 11q23 as does 22q11 during meiosis, chromosome 21 rarely rearranges with 11q23, and the DNA sequence of chromosome 21 appears to be less susceptible than 22q11 to double-strand breaks (DSBs). It has been suggested that the rearrangement recurs as a result of the palindromic AT-rich repeats at both 11q23 and 22q11, which extrude hairpin structures that are susceptible to DSBs. To determine whether the DSBs at these sites coincide with normal hotspots of meiotic recombination, immunocytochemical mapping of MLH1, a protein involved in crossing over, was employed. The results indicate that the translocation breakpoints do not coincide with recombination hotspots and therefore are unlikely to be the result of meiotic programmed DSBs, although MRE11 is likely to be involved. Previous analysis indicated that the DSBs appear to be repaired by a mechanism similar to nonhomologous end joining (NHEJ), although NHEJ is normally suppressed during meiosis. Taken together, these studies support the hypothesis that physical proximity between 11q23 and 22q11--but not typical meiotic recombinational activity in meiotic prophase--plays an important role in the generation of the constitutional t(11;22) rearrangement.
Figures



References
Web Resources
-
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for BAC 442e11 [accession number AC007707], PATRR11 [accession number AF391129], PATRR17 [accession number AB195814], and PATRR22 [accession numbers AC087065 and AC074203])
-
- MicroMeasure, http://www.colostate.edu/Depts/Biology/MicroMeasure
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for Emanuel syndrome)
-
- PALINDROME, http://bioweb.pasteur.fr/seqanal/interfaces/palindrome.html (for the EMBOSS palindrome recognition program)
References
-
- Fraccaro M, Lindsten J, Ford C, Iselius L (1980) The 11q;22q translocation: a European collaborative analysis of 43 cases. Hum Genet 56:21–51 - PubMed
-
- Lin TY, Viswanathan S, Wood C, Wilson PG, Wolf N, Fuller MT (1996) Coordinate developmental control of the meiotic cell cycle and spermatid defferentiation in Drosophila males. Development 122:1331–1341 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

