A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced
- PMID: 16912274
- PMCID: PMC1553206
- DOI: 10.1101/gad.380906
A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced
Abstract
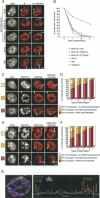
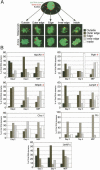
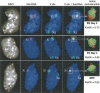
During early mammalian female development, one of the two X chromosomes becomes inactivated. Although X-chromosome coating by Xist RNA is essential for the initiation of X inactivation, little is known about how this signal is transformed into transcriptional silencing. Here we show that exclusion of RNA Polymerase II and transcription factors from the Xist RNA-coated X chromosome represents the earliest event following Xist RNA accumulation described so far in differentiating embryonic stem (ES) cells. Paradoxically, exclusion of the transcription machinery occurs before gene silencing is complete. However, examination of the three-dimensional organization of X-linked genes reveals that, when transcribed, they are always located at the periphery of, or outside, the Xist RNA domain, in contact with the transcription machinery. Upon silencing, genes shift to a more internal location, within the Xist RNA compartment devoid of transcription factors. Surprisingly, the appearance of this compartment is not dependent on the A-repeats of the Xist transcript, which are essential for gene silencing. However, the A-repeats are required for the relocation of genes into the Xist RNA silent domain. We propose that Xist RNA has multiple functions: A-repeat-independent creation of a transcriptionally silent nuclear compartment; and A-repeat-dependent induction of gene repression, which is associated with their translocation into this silent domain.
Figures



References
-
- Avner P., Heard E. X-chromosome inactivation: Counting, choice and initiation. Nat. Rev. Genet. 2001;2:59–67. - PubMed
-
- Boggs B.A., Connors B., Sobel R.E., Chinault A.C., Allis C.D. Reduced levels of histone H3 acetylation on the inactive X chromosome in human females. Chromosoma. 1996;105:303–309. - PubMed
-
- Boggs B.A., Cheung P., Heard E., Spector D.L., Chinault A.C., Allis C.D. Differentially methylated forms of his-tone H3 show unique association patterns with inactive human X chromosomes. Nat. Genet. 2002;30:73–76. - PubMed
-
- Brown C.J., Hendrich B.D., Rupert J.L., Lafreniere R.G., Xing Y., Lawrence J., Willard H.F. The human XIST gene: Analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
