Plasticity and expanding complexity of the hepatic transcription factor network during liver development
- PMID: 16912278
- PMCID: PMC1553211
- DOI: 10.1101/gad.390906
Plasticity and expanding complexity of the hepatic transcription factor network during liver development
Abstract
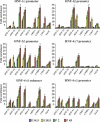
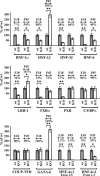
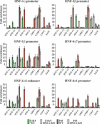
Cross-regulatory cascades between hepatic transcription factors have been implicated in the determination of the hepatic phenotype. Analysis of recruitments to regulatory regions and the temporal and spatial expression pattern of the main hepatic regulators during liver development revealed a gradual increase in complexity of autoregulatory and cross-regulatory circuits. Within these circuits we identified a core group of six transcription factors, which regulate the expression of each other and the expression of other downstream hepatic regulators. Changes in the promoter occupancy patterns during development included new recruitments, release, and exchange of specific factors. We also identified promoter and developmental stage-specific dual regulatory functions of certain factors as an important feature of the network. Inactivation of HNF-4alpha in embryonic, but not in adult, liver resulted in the diminished expression of most hepatic factors, demonstrating that the stability of the network correlates with its complexity. The results illustrate the remarkable flexibility of a self-sustaining transcription factor network, built up by complex dominant and redundant regulatory motifs in developing hepatocytes.
Figures




References
-
- Bailly A., Spath G., Bender V., Weiss M.C. Phenotypic effects of the forced expression of HNF4 and HNF1α are conditioned by properties of the recipient cell. J. Cell Sci. 1998;111:2411–2421. - PubMed
-
- Bossard P., Zaret K.S. Repressive and restrictive mesodermal interactions with gut endoderm: Possible relation to Meckel's Diverticulum. Development. 2000;127:4915–4923. - PubMed
-
- Briancon N., Bailly A., Clotman F., Jacquemin P., Lemaigre F.P., Weiss M.C. Expression of the α7 isoform of hepatocyte nuclear factor (HNF) 4 is activated by HNF6/ OC-2 and HNF1 and repressed by HNF4α1 in the liver. J. Biol. Chem. 2004;279:33398–33408. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
