Antiviral potency of APOBEC proteins does not correlate with cytidine deamination
- PMID: 16912295
- PMCID: PMC1563846
- DOI: 10.1128/JVI.00839-06
Antiviral potency of APOBEC proteins does not correlate with cytidine deamination
Abstract
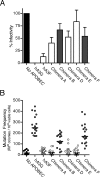
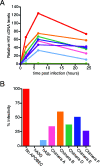
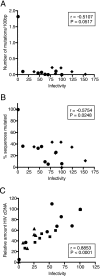
The human cytidine deaminases APOBEC3G (hA3G) and APOBEC3F (hA3F) are intracellular antiretroviral factors that can hypermutate nascent reverse transcripts and inhibit the replication of human immunodeficiency virus type 1 (HIV-1). Both enzymes have two cytidine deaminase motifs, although only the C-terminal motif is catalytic. Current models of APOBEC protein function imply editing is the principal mechanism of antiviral activity. In particular, hA3G is a more potent inhibitor of HIV-1 infectivity than hA3F and also induces a greater frequency of mutations in HIV-1 cDNA. We used hA3G/hA3F chimeric proteins to investigate whether cytidine deaminase potential reflects antiviral potency. We show here that the origin of the C-terminal deaminase motif is sufficient to determine the degree of mutation induced in a bacterial assay that measures mutations in chromosomal DNA. In contrast, this was not the case in the context of HIV-1 infection where the N-terminal deaminase motif also modulated the editing capabilities of the chimeras. Surprisingly, although three of the chimeric proteins induced levels of mutation that approximated those of parental hA3F, they displayed lower levels of antiviral activity. Most importantly, real-time PCR experiments revealed that the quantity of reverse transcripts detected in target cells, rather than the mutational burden carried by such DNAs, corresponded closely with viral infectivity. In other words, the antiviral phenotype of APOBEC proteins correlates with their ability to prevent the accumulation of reverse transcripts and not with the induction of hypermutation.
Figures


References
-
- Bishop, K. N., R. K. Holmes, A. M. Sheehy, N. O. Davidson, S. J. Cho, and M. H. Malim. 2004. Cytidine deamination of retroviral DNA by diverse. - PubMed
-
- Bishop, K. N., R. K. Holmes, A. M. Sheehy, and M. H. Malim. 2004. APOBEC-mediated editing of viral RNA. Science 305:645. - PubMed
-
- Chen, H., C. E. Lilley, Q. Yu, D. V. Lee, J. Chou, I. Narvaiza, N. R. Landau, and M. D. Weitzman. 2006. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr. Biol. 16:480-485. - PubMed
-
- Chiu, Y. L., V. B. Soros, J. F. Kreisberg, K. Stopak, W. Yonemoto, and W. C. Greene. 2005. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature 435:108-114. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

