Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains
- PMID: 16912299
- PMCID: PMC1563857
- DOI: 10.1128/JVI.00713-06
Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains
Abstract
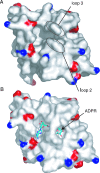
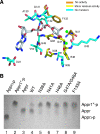
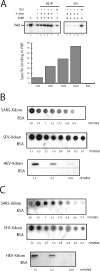
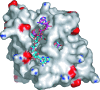
Macro domains constitute a protein module family found associated with specific histones and proteins involved in chromatin metabolism. In addition, a small number of animal RNA viruses, such as corona- and toroviruses, alphaviruses, and hepatitis E virus, encode macro domains for which, however, structural and functional information is extremely limited. Here, we characterized the macro domains from hepatitis E virus, Semliki Forest virus, and severe acute respiratory syndrome coronavirus (SARS-CoV). The crystal structure of the SARS-CoV macro domain was determined at 1.8-Angstroms resolution in complex with ADP-ribose. Information derived from structural, mutational, and sequence analyses suggests a close phylogenetic and, most probably, functional relationship between viral and cellular macro domain homologs. The data revealed that viral macro domains have relatively poor ADP-ribose 1"-phosphohydrolase activities (which were previously proposed to be their biologically relevant function) but bind efficiently free and poly(ADP-ribose) polymerase 1-bound poly(ADP-ribose) in vitro. Collectively, these results suggest to further evaluate the role of viral macro domains in host response to viral infection.
Figures



References
-
- Aguiar, R. C., K. Takeyama, C. He, K. Kreinbrink, and M. A. Shipp. 2005. B-aggressive lymphoma family proteins have unique domains that modulate transcription and exhibit poly(ADP-ribose) polymerase activity. J. Biol. Chem. 280:33756-33765. - PubMed
-
- Allen, M. D., A. M. Buckle, S. C. Cordell, J. Lowe, and M. Bycroft. 2003. The crystal structure of AF1521 a protein from Archaeoglobus fulgidus with homology to the non-histone domain of macroH2A. J. Mol. Biol. 330:503-511. - PubMed
-
- Cohen, S. X., R. J. Morris, F. J. Fernandez, M. Ben Jelloul, M. Kakaris, V. Parthasarathy, V. S. Lamzin, G. J. Kleywegt, and A. Perrakis. 2004. Towards complete validated models in the next generation of ARP/wARP. Acta Crystallogr. D Biol. Crystallogr. 60:2222-2229. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

