Tertiary structural and functional analyses of a viroid RNA motif by isostericity matrix and mutagenesis reveal its essential role in replication
- PMID: 16912306
- PMCID: PMC1563885
- DOI: 10.1128/JVI.00837-06
Tertiary structural and functional analyses of a viroid RNA motif by isostericity matrix and mutagenesis reveal its essential role in replication
Abstract
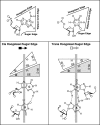
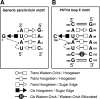
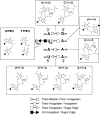
RNA-templated RNA replication is essential for viral or viroid infection, as well as for regulation of cellular gene expression. Specific RNA motifs likely regulate various aspects of this replication. Viroids of the Pospiviroidae family, as represented by the Potato spindle tuber viroid (PSTVd), replicate in the nucleus by utilizing DNA-dependent RNA polymerase II. We investigated the role of the loop E (sarcin/ricin) motif of the PSTVd genomic RNA in replication. A tertiary-structural model of this motif, inferred by comparative sequence analysis and comparison with nuclear magnetic resonance and X-ray crystal structures of loop E motifs in other RNAs, is presented in which core non-Watson-Crick base pairs are precisely specified. Isostericity matrix analysis of these base pairs showed that the model accounts for the reported natural sequence variations and viable experimental mutations in loop E motifs of PSTVd and other viroids. Furthermore, isostericity matrix analysis allowed us to design disruptive, as well as compensatory, mutations of PSTVd loop E. Functional analyses of such mutants by in vitro and in vivo experiments demonstrated that loop E structural integrity is crucial for replication, specifically during transcription. Our results suggest that the PSTVd loop E motif exists and functions in vivo and provide loss-of-function genetic evidence for the essential role of a viroid RNA three-dimensional motif in rolling-circle replication. The use of isostericity matrix analysis of non-Watson-Crick base pairing to rationalize mutagenesis of tertiary motifs and systematic in vitro and in vivo functional assays of mutants offers a novel, comprehensive approach to elucidate the tertiary-structure-function relationships for RNA motifs of general biological significance.
Figures

Similar articles
-
Existence in vivo of the loop E motif in potato spindle tuber viroid RNA.Arch Virol. 2007;152(7):1389-93. doi: 10.1007/s00705-007-0952-y. Epub 2007 Mar 19. Arch Virol. 2007. PMID: 17370107
-
A mini-RNA containing the tetraloop, wobble-pair and loop E motifs of the central conserved region of potato spindle tuber viroid is processed into a minicircle.Nucleic Acids Res. 2003 Feb 1;31(3):988-98. doi: 10.1093/nar/gkg193. Nucleic Acids Res. 2003. PMID: 12560495 Free PMC article.
-
A three-dimensional RNA motif in Potato spindle tuber viroid mediates trafficking from palisade mesophyll to spongy mesophyll in Nicotiana benthamiana.Plant Cell. 2011 Jan;23(1):258-72. doi: 10.1105/tpc.110.081414. Epub 2011 Jan 21. Plant Cell. 2011. PMID: 21258006 Free PMC article.
-
Potato Spindle Tuber Viroid RNA-Templated Transcription: Factors and Regulation.Viruses. 2018 Sep 17;10(9):503. doi: 10.3390/v10090503. Viruses. 2018. PMID: 30227597 Free PMC article. Review.
-
Viroids: unusual small pathogenic RNAs.Acta Biochim Pol. 2004;51(3):587-607. Acta Biochim Pol. 2004. PMID: 15448723 Review.
Cited by
-
Thermostable RNase P RNAs lacking P18 identified in the Aquificales.RNA. 2006 Nov;12(11):1915-21. doi: 10.1261/rna.242806. Epub 2006 Sep 27. RNA. 2006. PMID: 17005927 Free PMC article.
-
Viroid pathogenicity: one process, many faces.Viruses. 2009 Sep;1(2):298-316. doi: 10.3390/v1020298. Epub 2009 Sep 10. Viruses. 2009. PMID: 21994551 Free PMC article.
-
The transcription initiation sites of eggplant latent viroid strands map within distinct motifs in their in vivo RNA conformations.RNA Biol. 2016;13(1):83-97. doi: 10.1080/15476286.2015.1119365. RNA Biol. 2016. PMID: 26618399 Free PMC article.
-
A three-dimensional RNA motif mediates directional trafficking of Potato spindle tuber viroid from epidermal to palisade mesophyll cells in Nicotiana benthamiana.PLoS Pathog. 2019 Oct 23;15(10):e1008147. doi: 10.1371/journal.ppat.1008147. eCollection 2019 Oct. PLoS Pathog. 2019. PMID: 31644572 Free PMC article.
-
RNA three-dimensional structure drives the sequence organization of potato spindle tuber viroid quasispecies.PLoS Pathog. 2024 Apr 4;20(4):e1012142. doi: 10.1371/journal.ppat.1012142. eCollection 2024 Apr. PLoS Pathog. 2024. PMID: 38574111 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

