Marek's disease virus encodes MicroRNAs that map to meq and the latency-associated transcript
- PMID: 16912324
- PMCID: PMC1563840
- DOI: 10.1128/JVI.00831-06
Marek's disease virus encodes MicroRNAs that map to meq and the latency-associated transcript
Abstract
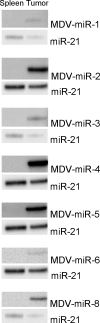
MicroRNAs (miRNAs) are a class of small (approximately 22-nucleotide) regulatory molecules that block translation or induce degradation of target mRNAs. These have been identified in a wide range of organisms, including viruses. In particular, the oncogenic gammaherpesviruses Kaposi's sarcoma herpesvirus and Epstein-Barr virus encode miRNAs that could potentially regulate either viral or host genes. To determine if Marek's disease virus (MDV), an oncogenic alphaherpesvirus of chickens, encodes miRNAs, we isolated small RNAs from MDV-infected chicken embryo fibroblasts (CEF) and used the 454 Life Sciences sequencing technology to obtain the sequences of 13,679 candidate host and viral small RNAs. Eight miRNAs were found, five of which flank the meq oncogene and three that map to the latency-associated transcript (LAT) region of the genome. The meq gene is unique to pathogenic serotypes of MDV and is transcriptionally active during latency and transformation, and the LAT region of the MDV genome is antisense to the immediate-early gene ICP4. Secondary structure analysis predicted that the regions flanking the miRNAs could form hairpin precursors. Northern blot analysis confirmed expression of all miRNAs in MDV-infected CEF, MDV-induced tumors, and MDV lymphoblastoid cell lines. We propose that the MDV miRNAs function to enable MDV pathogenesis and contribute to MDV-induced transformation of chicken T cells.
Figures




References
-
- Akiyama, Y., S. Kato, and N. Iwa. 1973. Continuous cell culture from lymphoma of Marek's disease. Biken J. 16:177-179. - PubMed
-
- Aravin, A. A., M. Lagos-Quintana, A. Yalcin, M. Zavolan, D. Marks, B. Snyder, T. Gaasterland, J. Meyer, and T. Tuschl. 2003. The small RNA profile during Drosophila melanogaster development. Dev. Cell 5:337-350. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

