Gene function correlates with potential for G4 DNA formation in the human genome
- PMID: 16914419
- PMCID: PMC1557811
- DOI: 10.1093/nar/gkl529
Gene function correlates with potential for G4 DNA formation in the human genome
Abstract
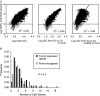
G-rich genomic regions can form G4 DNA upon transcription or replication. We have quantified the potential for G4 DNA formation (G4P) of the 16 654 genes in the human RefSeq database, and then correlated gene function with G4P. We have found that very low and very high G4P correlates with specific functional classes of genes. Notably, tumor suppressor genes have very low G4P and proto-oncogenes have very high G4P. G4P of these genes is evenly distributed between exons and introns, and it does not reflect enrichment for CpG islands or local chromosomal environment. These results show that genomic structure undergoes selection based on gene function. Selection based on G4P could promote genomic stability (or instability) of specific classes of genes; or reflect mechanisms for global regulation of gene expression.
Figures




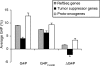
References
-
- Sen D., Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

