Comprehensive splice-site analysis using comparative genomics
- PMID: 16914448
- PMCID: PMC1557818
- DOI: 10.1093/nar/gkl556
Comprehensive splice-site analysis using comparative genomics
Abstract
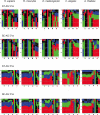
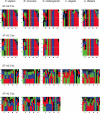
We have collected over half a million splice sites from five species-Homo sapiens, Mus musculus, Drosophila melanogaster, Caenorhabditis elegans and Arabidopsis thaliana-and classified them into four subtypes: U2-type GT-AG and GC-AG and U12-type GT-AG and AT-AC. We have also found new examples of rare splice-site categories, such as U12-type introns without canonical borders, and U2-dependent AT-AC introns. The splice-site sequences and several tools to explore them are available on a public website (SpliceRack). For the U12-type introns, we find several features conserved across species, as well as a clustering of these introns on genes. Using the information content of the splice-site motifs, and the phylogenetic distance between them, we identify: (i) a higher degree of conservation in the exonic portion of the U2-type splice sites in more complex organisms; (ii) conservation of exonic nucleotides for U12-type splice sites; (iii) divergent evolution of C.elegans 3' splice sites (3'ss) and (iv) distinct evolutionary histories of 5' and 3'ss. Our study proves that the identification of broad patterns in naturally-occurring splice sites, through the analysis of genomic datasets, provides mechanistic and evolutionary insights into pre-mRNA splicing.
Figures




References
-
- Collins L., Penny D. Complex spliceosomal organization ancestral to extant eukaryotes. Mol. Biol. Evol. 2005;22:1053–1066. - PubMed
-
- Brow D.A. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 2002;36:333–360. - PubMed
-
- Hastings M.L., Krainer A.R. Pre-mRNA splicing in the new millennium. Curr. Opin. Cell. Biol. 2001;13:302–309. - PubMed
-
- Will C.L., Luhrmann R. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell. Biol. 2001;13:290–301. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

