Dynamic regulation of Drosophila nuclear receptor activity in vivo
- PMID: 16914501
- PMCID: PMC2100403
- DOI: 10.1242/dev.02512
Dynamic regulation of Drosophila nuclear receptor activity in vivo
Abstract
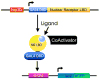
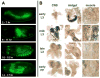
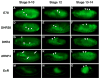
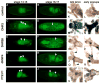
Nuclear receptors are a large family of transcription factors that play major roles in development, metamorphosis, metabolism and disease. To determine how, where and when nuclear receptors are regulated by small chemical ligands and/or protein partners, we have used a 'ligand sensor' system to visualize spatial activity patterns for each of the 18 Drosophila nuclear receptors in live developing animals. Transgenic lines were established that express the ligand binding domain of each nuclear receptor fused to the DNA-binding domain of yeast GAL4. When combined with a GAL4-responsive reporter gene, the fusion proteins show tissue- and stage-specific patterns of activation. We show that these responses accurately reflect the presence of endogenous and exogenously added hormone, and that they can be modulated by nuclear receptor partner proteins. The amnioserosa, yolk, midgut and fat body, which play major roles in lipid storage, metabolism and developmental timing, were identified as frequent sites of nuclear receptor activity. We also see dynamic changes in activation that are indicative of sweeping changes in ligand and/or co-factor production. The screening of a small compound library using this system identified the angular psoralen angelicin and the insect growth regulator fenoxycarb as activators of the Ultraspiracle (USP) ligand-binding domain. These results demonstrate the utility of this system for the functional dissection of nuclear receptor pathways and for the development of new receptor agonists and antagonists that can be used to modulate metabolism and disease and to develop more effective means of insect control.
Figures



References
-
- Agoulnik IU, Tong XW, Fischer DC, Korner K, Atkinson NE, Edwards DP, Headon DR, Weigel NL, Kieback DG. A germline variation in the progesterone receptor gene increases transcriptional activity and may modify ovarian cancer risk. J Clin Endocrinol Metab. 2004;89:6340–6347. - PubMed
-
- Andres AJ, Thummel CS. Methods for quantitative analysis of transcription in larvae and prepupae. Methods Cell Biol. 1994;44:565–573. - PubMed
-
- Andres AJ, Fletcher JC, Karim FD, Thummel CS. Molecular analysis of the initiation of insect metamorphosis: a comparative study of Drosophila ecdysteroid-regulated transcription. Dev Biol. 1993;160:388–404. - PubMed
-
- Ashburner M. Patterns of puffing activity in the salivary gland chromosomes of Drosophila. VI Induction by ecdysone in salivary glands of D melanogaster cultured in vitro. Chromosoma. 1972;38:255–281. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

