Activation of receptor activator of NF-kappaB ligand gene expression by 1,25-dihydroxyvitamin D3 is mediated through multiple long-range enhancers
- PMID: 16914732
- PMCID: PMC1592822
- DOI: 10.1128/MCB.00353-06
Activation of receptor activator of NF-kappaB ligand gene expression by 1,25-dihydroxyvitamin D3 is mediated through multiple long-range enhancers
Abstract
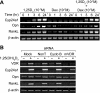
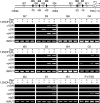
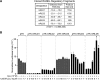
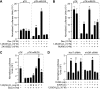
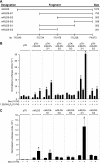
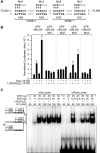
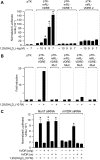
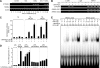
RANKL is a tumor necrosis factor (TNF)-like factor secreted by mesenchymal cells, osteoblast derivatives, and T cells that is essential for osteoclastogenesis. In osteoblasts, RANKL expression is regulated by two major calcemic hormones, 1,25-dihydroxyvitamin D(3) [1,25(OH)(2)D(3)] and parathyroid hormone (PTH), as well as by several inflammatory/osteoclastogenic cytokines; the molecular mechanisms for this regulation are unclear. To identify such mechanisms, we screened a DNA microarray which tiled across the entire mouse RankL gene locus at a 50-bp resolution using chromatin immunoprecipitation (ChIP)-derived DNA precipitated with antibodies to the vitamin D receptor (VDR) and the retinoid X receptor (RXR). Five sites of dimer interaction were observed on the RankL gene centered at 16, 22, 60, 69, and 76 kb upstream of the TSS. These regions contained binding sites for not only VDR and RXR, but also the glucocorticoid receptor (GR). The most distant of these regions, termed the distal control region (RL-DCR), conferred both VDR-dependent 1,25(OH)(2)D(3) and GR-dependent glucocorticoid (GC) responses. We mapped these activities to an unusual but functionally active vitamin D response element and to several potential GC response elements located over a more extensive region within the RL-DCR. An evolutionarily conserved region within the human RANKL gene contained a similar vitamin D response element and exhibited an equivalent behavior. Importantly, hormonal activation of the RankL gene was also associated with chromatin modification and RNA polymerase II recruitment. Our studies demonstrate that regulation of RankL gene expression by 1,25(OH)(2)D(3) is complex and mediated by at least five distal regions, one of which contains a specific element capable of mediating direct transcriptional activation.
Figures


References
-
- Bettoun, D. J., T. P. Burris, K. A. Houck, D. W. Buck II, K. R. Stayrook, B. Khalifa, J. Lu, W. W. Chin, and S. Nagpal. 2003. Retinoid X receptor is a nonsilent major contributor to vitamin D receptor-mediated transcriptional activation. Mol. Endocrinol. 17:2320-2328. [Epub ahead of print.] - PubMed
-
- Carroll, J. S., X. S. Liu, A. S. Brodsky, W. Li, C. A. Meyer, A. J. Szary, J. Eeckhoute, W. Shao, E. V. Hestermann, T. R. Geistlinger, E. A. Fox, P. A. Silver, and M. Brown. 2005. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122:33-43. - PubMed
-
- Cawley, S., S. Bekiranov, H. H. Ng, P. Kapranov, E. A. Sekinger, D. Kampa, A. Piccolboni, V. Sementchenko, J. Cheng, A. J. Williams, R. Wheeler, B. Wong, J. Drenkow, M. Yamanaka, S. Patel, S. Brubaker, H. Tammana, G. Helt, K. Struhl, and T. R. Gingeras. 2004. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 116:499-509. - PubMed
-
- Cheng, J., P. Kapranov, J. Drenkow, S. Dike, S. Brubaker, S. Patel, J. Long, D. Stern, H. Tammana, G. Helt, V. Sementchenko, A. Piccolboni, S. Bekiranov, D. K. Bailey, M. Ganesh, S. Ghosh, I. Bell, D. S. Gerhard, and T. R. Gingeras. 2005. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 308:1149-1154. [Epub ahead of print.] - PubMed
-
- Cosgrove, M. S., J. D. Boeke, and C. Wolberger. 2004. Regulated nucleosome mobility and the histone code. Nat. Struct. Mol. Biol. 11:1037-1043. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
