Integration of metabolite with transcript and enzyme activity profiling during diurnal cycles in Arabidopsis rosettes
- PMID: 16916443
- PMCID: PMC1779593
- DOI: 10.1186/gb-2006-7-8-R76
Integration of metabolite with transcript and enzyme activity profiling during diurnal cycles in Arabidopsis rosettes
Abstract
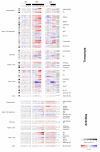
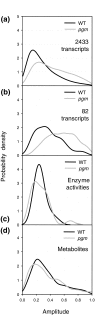
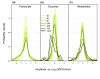
Background: Genome-wide transcript profiling and analyses of enzyme activities from central carbon and nitrogen metabolism show that transcript levels undergo marked and rapid changes during diurnal cycles and after transfer to darkness, whereas changes in activities are smaller and delayed. In the starchless pgm mutant, where sugars are depleted every night, there are accentuated diurnal changes in transcript levels. Enzyme activities in this mutant do not show larger diurnal changes; instead, they shift towards the levels found in the wild type after several days of darkness. This indicates that enzyme activities change slowly, integrating the changes in transcript levels over several diurnal cycles.
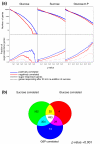
Results: To generalize this conclusion, 137 metabolites were profiled using gas and liquid chromatography coupled to mass spectroscopy. The amplitudes of the diurnal changes in metabolite levels in pgm were (with the exception of sugars) similar or smaller than in the wild type. The average levels shifted towards those found after several days of darkness in the wild type. Examples include increased levels of amino acids due to protein degradation, decreased levels of fatty acids, increased tocopherol and decreased myo-inositol. Many metabolite-transcript correlations were found and the proportion of transcripts correlated with sugars increased dramatically in the starchless mutant.
Conclusion: Rapid diurnal changes in transcript levels are integrated over time to generate quasi-stable changes across large sectors of metabolism. This implies that correlations between metabolites and transcripts are due to regulation of gene expression by metabolites, rather than metabolites being changed as a consequence of a change in gene expression.
Figures







Similar articles
-
A Robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness.Plant Cell. 2004 Dec;16(12):3304-25. doi: 10.1105/tpc.104.025973. Epub 2004 Nov 17. Plant Cell. 2004. PMID: 15548738 Free PMC article.
-
Temporal responses of transcripts, enzyme activities and metabolites after adding sucrose to carbon-deprived Arabidopsis seedlings.Plant J. 2007 Feb;49(3):463-91. doi: 10.1111/j.1365-313X.2006.02979.x. Epub 2007 Jan 1. Plant J. 2007. PMID: 17217462
-
Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis.Plant Cell. 2005 Dec;17(12):3257-81. doi: 10.1105/tpc.105.035261. Epub 2005 Nov 18. Plant Cell. 2005. PMID: 16299223 Free PMC article.
-
Steps towards an integrated view of nitrogen metabolism.J Exp Bot. 2002 Apr;53(370):959-70. doi: 10.1093/jexbot/53.370.959. J Exp Bot. 2002. PMID: 11912238 Review.
-
A network perspective on nitrogen metabolism from model to crop plants using integrated 'omics' approaches.J Exp Bot. 2014 Oct;65(19):5619-30. doi: 10.1093/jxb/eru322. Epub 2014 Aug 16. J Exp Bot. 2014. PMID: 25129130 Review.
Cited by
-
uORFs: Important Cis-Regulatory Elements in Plants.Int J Mol Sci. 2020 Aug 28;21(17):6238. doi: 10.3390/ijms21176238. Int J Mol Sci. 2020. PMID: 32872304 Free PMC article. Review.
-
Dynamic plastid redox signals integrate gene expression and metabolism to induce distinct metabolic states in photosynthetic acclimation in Arabidopsis.Plant Cell. 2009 Sep;21(9):2715-32. doi: 10.1105/tpc.108.062018. Epub 2009 Sep 8. Plant Cell. 2009. PMID: 19737978 Free PMC article.
-
An L,L-diaminopimelate aminotransferase mutation leads to metabolic shifts and growth inhibition in Arabidopsis.J Exp Bot. 2018 Nov 26;69(22):5489-5506. doi: 10.1093/jxb/ery325. J Exp Bot. 2018. PMID: 30215754 Free PMC article.
-
Enzyme activity profiles during fruit development in tomato cultivars and Solanum pennellii.Plant Physiol. 2010 May;153(1):80-98. doi: 10.1104/pp.110.154336. Epub 2010 Mar 24. Plant Physiol. 2010. PMID: 20335402 Free PMC article.
-
Data Reduction Approaches for Dissecting Transcriptional Effects on Metabolism.Front Plant Sci. 2018 Apr 20;9:538. doi: 10.3389/fpls.2018.00538. eCollection 2018. Front Plant Sci. 2018. PMID: 29731765 Free PMC article.
References
-
- Stitt M, Sonnewald U. Regulation of metabolism in transgenic plants. Annu Rev Plant Biol. 1995;46:341–361. doi: 10.1146/annurev.pp.46.060195.002013. - DOI
-
- Gibon Y, Blaesing OE, Hannemann J, Carillo P, Hoehne M, Hendriks JHM, Palacios N, Cross J, Selbig J, Stitt M. A Robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell. 2004;16:3304–3325. doi: 10.1105/tpc.104.025973. - DOI - PMC - PubMed
-
- Hirai MY, Yano M, Goodenowe DB, Kanaya S, Kimura T, Awazuhara M, Arita M, Fujiwara T, Saito K. Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2004;101:10205–10210. doi: 10.1073/pnas.0403218101. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

