The N-CoR complex enables chromatin remodeler SNF2H to enhance repression by thyroid hormone receptor
- PMID: 16917504
- PMCID: PMC1560369
- DOI: 10.1038/sj.emboj.7601280
The N-CoR complex enables chromatin remodeler SNF2H to enhance repression by thyroid hormone receptor
Abstract
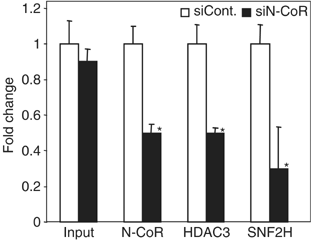
Unliganded thyroid hormone receptor (TR) actively represses transcription via the nuclear receptor corepressor (N-CoR)/histone deacetylase 3 (HDAC3) complex. Although transcriptional activation by liganded receptors involves chromatin remodeling, the role of ATP-dependent remodeling in receptor-mediated repression is unknown. Here we report that SNF2H, the mammalian ISWI chromatin remodeling ATPase, is critical for repression of a genomically integrated, TR-regulated reporter gene. N-CoR and HDAC3 are both required for recruitment of SNF2H to the repressed gene. SNF2H does not interact directly with the N-CoR/HDAC3 complex, but binds to unacetylated histone H4 tails, suggesting that deacetylase activity of the corepressor complex is critical to SNF2H function. Indeed, HDAC3 as well as SNF2H are required for nucleosomal organization on the TR target gene. Consistent with these findings, reduction of SNF2H induces expression of an endogenous TR-regulated gene, dio1, in liver cells. Thus, although not apparent from studies of transiently transfected reporter genes, gene repression by TR involves the targeting of chromatin remodeling factors to repressed genes by the HDAC activity of nuclear receptor corepressors.
Figures








Similar articles
-
The N-CoR/histone deacetylase 3 complex is required for repression by thyroid hormone receptor.Mol Cell Biol. 2003 Aug;23(15):5122-31. doi: 10.1128/MCB.23.15.5122-5131.2003. Mol Cell Biol. 2003. PMID: 12861000 Free PMC article.
-
Human THAP7 is a chromatin-associated, histone tail-binding protein that represses transcription via recruitment of HDAC3 and nuclear hormone receptor corepressor.J Biol Chem. 2005 Feb 25;280(8):7346-58. doi: 10.1074/jbc.M411675200. Epub 2004 Nov 23. J Biol Chem. 2005. PMID: 15561719
-
Targeting of N-CoR and histone deacetylase 3 by the oncoprotein v-erbA yields a chromatin infrastructure-dependent transcriptional repression pathway.EMBO J. 2000 Aug 1;19(15):4074-90. doi: 10.1093/emboj/19.15.4074. EMBO J. 2000. PMID: 10921888 Free PMC article.
-
N-CoR-HDAC corepressor complexes: roles in transcriptional regulation by nuclear hormone receptors.Curr Top Microbiol Immunol. 2003;274:237-68. doi: 10.1007/978-3-642-55747-7_9. Curr Top Microbiol Immunol. 2003. PMID: 12596910 Review.
-
Modulation of thyroid hormone receptor silencing function by co-repressors and a synergizing transcription factor.Biochem Soc Trans. 2000;28(4):386-9. Biochem Soc Trans. 2000. PMID: 10961925 Review.
Cited by
-
A Role for the PPARgamma in Cancer Therapy.PPAR Res. 2008;2008:314974. doi: 10.1155/2008/314974. PPAR Res. 2008. PMID: 18528521 Free PMC article.
-
The Role of Histone Deacetylase 3 Complex in Nuclear Hormone Receptor Action.Int J Mol Sci. 2021 Aug 24;22(17):9138. doi: 10.3390/ijms22179138. Int J Mol Sci. 2021. PMID: 34502048 Free PMC article. Review.
-
Identification and characterization of a selective peroxisome proliferator-activated receptor beta/delta (NR1C2) antagonist.Mol Endocrinol. 2008 Feb;22(2):523-9. doi: 10.1210/me.2007-0190. Epub 2007 Nov 1. Mol Endocrinol. 2008. PMID: 17975020 Free PMC article.
-
Overlapping chromatin-remodeling systems collaborate genome wide at dynamic chromatin transitions.Nat Struct Mol Biol. 2014 Jan;21(1):73-81. doi: 10.1038/nsmb.2718. Epub 2013 Dec 8. Nat Struct Mol Biol. 2014. PMID: 24317492 Free PMC article.
-
Human ISWI complexes are targeted by SMARCA5 ATPase and SLIDE domains to help resolve lesion-stalled transcription.Nucleic Acids Res. 2014 Jul;42(13):8473-85. doi: 10.1093/nar/gku565. Epub 2014 Jul 2. Nucleic Acids Res. 2014. PMID: 24990377 Free PMC article.
References
-
- Aalfs JD, Narlikar GJ, Kingston RE (2001) Functional differences between the human ATP-dependent nucleosome remodeling proteins BRG1 and SNF2H. J Biol Chem 276: 34270–34278 - PubMed
-
- Aranda A, Pascual A (2001) Nuclear hormone receptors and gene expression. Physiol Rev 81: 1269–1304 - PubMed
-
- Berry MJ, Banu L, Larsen PR (1991) Type I iodothyronine deiodinase is a selenocysteine-containing enzyme. Nature 349: 438–440 - PubMed
-
- Cervoni N, Szyf M (2001) Demethylase activity is directed by histone acetylation. J Biol Chem 276: 40778–40787 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

