Breast cancer metastasis suppressor 1 (BRMS1) is stabilized by the Hsp90 chaperone
- PMID: 16919237
- PMCID: PMC1557677
- DOI: 10.1016/j.bbrc.2006.08.005
Breast cancer metastasis suppressor 1 (BRMS1) is stabilized by the Hsp90 chaperone
Abstract
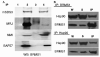
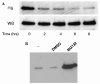
Breast cancer metastasis suppressor 1 (BRMS1) is a member of the mSin3-HDAC transcription co-repressor complex. However, the proteins associated with BRMS1 have not been fully identified. Yeast two-hybrid screen, immuno-affinity chromatography, and co-immunoprecipitation experiments were performed to identify BRMS1 interacting proteins (BIPs). In addition to known core mSin3 transcriptional complex components RBBP1 and mSDS3, BRMS1 interacted with other proteins including three chaperones: DNAJB6 (MRJ), Hsp90, and Hsp70. Hsp90 is a known target of HDAC6 and reversible acetylation is one of the mechanisms that is implicated in regulation of Hsp90 chaperone complex activity. BRMS1 interacted with class II HDACs, HDAC 4, 5, and 6. We further found that BRMS1 is stabilized by Hsp90, and its turnover is proteasome dependent. The stability of BRMS1 protein may be important in maintaining the functional role of BRMS1 in metastasis suppression.
Figures



Similar articles
-
Breast cancer metastasis suppressor 1 (BRMS1) forms complexes with retinoblastoma-binding protein 1 (RBP1) and the mSin3 histone deacetylase complex and represses transcription.J Biol Chem. 2004 Jan 9;279(2):1562-9. doi: 10.1074/jbc.M307969200. Epub 2003 Oct 26. J Biol Chem. 2004. PMID: 14581478
-
Breast cancer metastasis suppressor 1 (BRMS1) inhibits osteopontin transcription by abrogating NF-kappaB activation.Mol Cancer. 2007 Jan 16;6:6. doi: 10.1186/1476-4598-6-6. Mol Cancer. 2007. PMID: 17227585 Free PMC article.
-
Alterations of BRMS1-ARID4A interaction modify gene expression but still suppress metastasis in human breast cancer cells.J Biol Chem. 2008 Mar 21;283(12):7438-44. doi: 10.1074/jbc.M709446200. Epub 2008 Jan 22. J Biol Chem. 2008. PMID: 18211900 Free PMC article.
-
Drugging the HDAC6-HSP90 interplay in malignant cells.Trends Pharmacol Sci. 2014 Oct;35(10):501-9. doi: 10.1016/j.tips.2014.08.001. Epub 2014 Sep 16. Trends Pharmacol Sci. 2014. PMID: 25234862 Review.
-
Breast cancer metastasis suppressor 1: update.Clin Exp Metastasis. 2003;20(1):45-50. doi: 10.1023/a:1022542519586. Clin Exp Metastasis. 2003. PMID: 12650606 Review.
Cited by
-
HDAC6 and ovarian cancer.Int J Mol Sci. 2013 May 2;14(5):9514-35. doi: 10.3390/ijms14059514. Int J Mol Sci. 2013. PMID: 23644884 Free PMC article. Review.
-
MicroRNA signature distinguishes the degree of aggressiveness of papillary thyroid carcinoma.Ann Surg Oncol. 2011 Jul;18(7):2035-41. doi: 10.1245/s10434-011-1733-0. Epub 2011 May 3. Ann Surg Oncol. 2011. PMID: 21537871 Free PMC article.
-
Novobiocin and additional inhibitors of the Hsp90 C-terminal nucleotide-binding pocket.Curr Med Chem. 2008;15(26):2702-17. doi: 10.2174/092986708786242895. Curr Med Chem. 2008. PMID: 18991631 Free PMC article. Review.
-
Expression of the Breast Cancer Metastasis Suppressor 1 (BRMS1) maintains in vitro chemosensitivity of breast cancer cells.Cancer Lett. 2009 Aug 18;281(1):100-7. doi: 10.1016/j.canlet.2009.02.035. Cancer Lett. 2009. PMID: 19307053 Free PMC article.
-
Cytoplasmic BRMS1 expression in malignant melanoma is associated with increased disease-free survival.BMC Cancer. 2012 Feb 22;12:73. doi: 10.1186/1471-2407-12-73. BMC Cancer. 2012. PMID: 22356677 Free PMC article.
References
-
- Kang YB, Siegel PM, Shu WP, Drobnjak M, Kakonen SM, Cordón-Cardo C, Guise TA, Massagué J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. - PubMed
-
- Debies MT, Welch DR. Genetic basis of human breast cancer metastasis. J. Mammary Gland Biol. Neoplasia. 2001;6:441–451. - PubMed
-
- Seraj MJ, Samant RS, Verderame MF, Welch DR. Functional evidence for a novel human breast carcinoma metastasis suppressor, BRMS1, encoded at chromosome 11q13. Cancer Res. 2000;60:2764–2769. - PubMed
-
- Samant RS, Seraj MJ, Saunders MM, Sakamaki T, Shevde LA, Harms JF, Leonard TO, Goldberg SF, Budgeon LR, Meehan WJ, Winter CR, Christensen ND, Verderame MF, Donahue HJ, Welch DR. Analysis of mechanisms underlying BRMS1 suppression of metastasis. Clin. Exp. Metastasis. 2001;18:683–693. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

