B29 gene silencing in pituitary cells is regulated by its 3' enhancer
- PMID: 16920149
- PMCID: PMC2104784
- DOI: 10.1016/j.jmb.2006.07.046
B29 gene silencing in pituitary cells is regulated by its 3' enhancer
Abstract
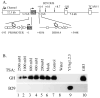
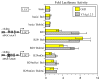
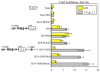
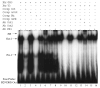
B cell-specific B29 (Igbeta, CD79b) genes in rat, mouse, and human are situated between the 5' growth hormone (GH) locus control region and the 3' GH gene cluster. The entire GH genomic region is DNase 1 hypersensitive in GH-expressing pituitary cells, which predicts an "open" chromatin configuration, and yet B29 is not expressed. The B29 promoter and enhancers exhibit histone deacetylation in pituitary cells, but histone deacetylase inhibition failed to activate B29 expression. The B29 promoter and a 3' enhancer showed local dense DNA methylation in both pituitary and non-lymphoid cells consistent with gene silencing. However, DNA methyltransferase inhibition did not activate B29 expression either. B29 promoter constructs were minimally activated in transfected pituitary cells. Co-transfection of the B cell-specific octamer transcriptional co-activator Bob1 with the B29 promoter construct resulted in high level promoter activity in pituitary cells comparable to B29 promoter activity in transfected B cells. Unexpectedly, inclusion of the B29 3' enhancer in B29 promoter constructs strongly inhibited B29 transcriptional activity even when pituitary cells were co-transfected with Bob1. Both Oct-1 and Pit-1 bind the B29 3' enhancer in in vitro electrophoretic mobility shift assay and in in vivo chromatin immunoprecipitation analyses. These data indicate that the GH locus-embedded, tissue-specific B29 gene is silenced in GH-expressing pituitary cells by epigenetic mechanisms, the lack of a B cell-specific transcription factor, and likely by the B29 3' enhancer acting as a powerful silencer in a context and tissue-specific manner.
Figures




Similar articles
-
A conserved sequence upstream of the B29 (Ig beta, CD79b) gene interacts with YY1.Mol Biol Rep. 2004 Mar;31(1):1-11. doi: 10.1023/b:mole.0000013489.04734.5e. Mol Biol Rep. 2004. PMID: 15040449
-
Bob1 (OCA-B/OBF-1) differential transactivation of the B cell-specific B29 (Ig beta) and mb-1 (Ig alpha) promoters.J Immunol. 2002 Apr 1;168(7):3369-75. doi: 10.4049/jimmunol.168.7.3369. J Immunol. 2002. PMID: 11907094
-
An upstream Oct-1- and Oct-2-binding silencer governs B29 (Ig beta) gene expression.J Immunol. 2000 Mar 1;164(5):2550-6. doi: 10.4049/jimmunol.164.5.2550. J Immunol. 2000. PMID: 10679093
-
Evidence for Pituitary Repression of the Human Growth Hormone-Related Placental Lactogen Genes and a Role for P Sequences.Int J Mol Sci. 2025 May 6;26(9):4421. doi: 10.3390/ijms26094421. Int J Mol Sci. 2025. PMID: 40362658 Free PMC article. Review.
-
Regulation of the human growth hormone gene family: possible role for Pit-1 in early stages of pituitary-specific expression and repression.Neuroendocrinology. 2006;83(3-4):145-53. doi: 10.1159/000095522. Epub 2006 Oct 13. Neuroendocrinology. 2006. PMID: 17047377 Review.
Cited by
-
Modulation of eIF5A expression using SNS01 nanoparticles inhibits NF-κB activity and tumor growth in murine models of multiple myeloma.Mol Ther. 2012 Jul;20(7):1305-14. doi: 10.1038/mt.2012.94. Epub 2012 May 15. Mol Ther. 2012. PMID: 22588272 Free PMC article.
-
Menadione Suppresses Benzo(α)pyrene-Induced Activation of Cytochromes P450 1A: Insights into a Possible Molecular Mechanism.PLoS One. 2016 May 11;11(5):e0155135. doi: 10.1371/journal.pone.0155135. eCollection 2016. PLoS One. 2016. PMID: 27167070 Free PMC article.
-
Maximal Expression of the Evolutionarily Conserved Slit2 Gene Promoter Requires Sp1.Cell Mol Neurobiol. 2016 Aug;36(6):955-964. doi: 10.1007/s10571-015-0281-8. Epub 2015 Oct 11. Cell Mol Neurobiol. 2016. PMID: 26456684 Free PMC article.
References
-
- Benschop RJ, Cambier JC. B cell development: signal transduction by antigen receptors and their surrogates. Curr Opin Immunol. 1999;11:143–51. - PubMed
-
- Thompson AA, Wood WJ, Jr, Gilly MJ, Damore MA, Omori SA, Wall R. The promoter and 5′ flanking sequences controlling human B29 gene expression. Blood. 1996;87:666–73. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

