Antisense-induced ribosomal frameshifting
- PMID: 16920740
- PMCID: PMC1616946
- DOI: 10.1093/nar/gkl531
Antisense-induced ribosomal frameshifting
Abstract
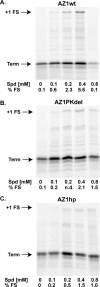
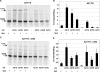
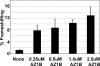
Programmed ribosomal frameshifting provides a mechanism to decode information located in two overlapping reading frames by diverting a proportion of translating ribosomes into a second open reading frame (ORF). The result is the production of two proteins: the product of standard translation from ORF1 and an ORF1-ORF2 fusion protein. Such programmed frameshifting is commonly utilized as a gene expression mechanism in viruses that infect eukaryotic cells and in a subset of cellular genes. RNA secondary structures, consisting of pseudoknots or stem-loops, located downstream of the shift site often act as cis-stimulators of frameshifting. Here, we demonstrate for the first time that antisense oligonucleotides can functionally mimic these RNA structures to induce +1 ribosomal frameshifting when annealed downstream of the frameshift site, UCC UGA. Antisense-induced shifting of the ribosome into the +1 reading frame is highly efficient in both rabbit reticulocyte lysate translation reactions and in cultured mammalian cells. The efficiency of antisense-induced frameshifting at this site is responsive to the sequence context 5' of the shift site and to polyamine levels.
Figures



References
-
- Baranov P.V., Gesteland R.F., Atkins J.F. Recoding: translational bifurcations in gene expression. Gene. 2002;286:187–201. - PubMed
-
- Namy O., Rousset J.P., Napthine S., Brierley I. Reprogrammed genetic decoding in cellular gene expression. Mol. Cell. 2004;13:157–168. - PubMed
-
- Gesteland R.F., Weiss R.B., Atkins J.F. Recoding: reprogrammed genetic decoding. Science. 1992;257:1640–1641. - PubMed
-
- Gesteland R.F., Atkins J.F. Recoding: dynamic reprogramming of translation. Annu. Rev. Biochem. 1996;65:741–768. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

