Mycobacterial transcriptional signals: requirements for recognition by RNA polymerase and optimal transcriptional activity
- PMID: 16920742
- PMCID: PMC1616969
- DOI: 10.1093/nar/gkl521
Mycobacterial transcriptional signals: requirements for recognition by RNA polymerase and optimal transcriptional activity
Abstract
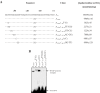
Majority of the promoter elements of mycobacteria do not function well in other eubacterial systems and analysis of their sequences has established the presence of only single conserved sequence located at the -10 position. Additional sequences for the appropriate functioning of these promoters have been proposed but not characterized, probably due to the absence of sufficient number of strong mycobacterial promoters. In the current study, we have isolated functional promoter-like sequences of mycobacteria from the pool of random DNA sequences. Based on the promoter activity in Mycobacterium smegmatis and score assigned by neural network promoter prediction program, we selected one of these promoter sequences, namely A37 for characterization in order to understand the structure of housekeeping promoters of mycobacteria. A37-RNAP complexes were subjected to DNase I footprinting and subsequent mutagenesis. Our results demonstrate that in addition to -10 sequences, DNA sequence at -35 site can also influence the activity of mycobacterial promoters by modulating the promoter recognition by RNA polymerase and subsequent formation of open complex. We also provide evidence that despite exhibiting similarities in -10 and -35 sequences, promoter regions of mycobacteria and Escherichia coli differ from each other due to differences in their requirement of spacer sequences between the two positions.
Figures




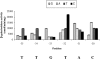

References
-
- Corbett E.L., Watt C.J., Walker N., Maher D., Williams B.G., Raviglione M.C., Dye C. The growing burden of tuberculosis: global trends and interactions with the HIV epidemic. Arch. Intern. Med. 2003;163:1009–1021. - PubMed
-
- Cole S.T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S.V., Eiglmeier K., Gas S., Barry C.E., III, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. - PubMed
-
- Sun R., Converse P.J., Ko C., Tyagi S., Morrison N.E., Bishai W.R. Mycobacterium tuberculosis ECF sigma factor sigC is required for lethality in mice and for the conditional expression of a defined gene set. Mol. Microbiol. 2004;52:25–38. - PubMed

