SUMO-conjugating and SUMO-deconjugating enzymes from Arabidopsis
- PMID: 16920872
- PMCID: PMC1557612
- DOI: 10.1104/pp.106.085415
SUMO-conjugating and SUMO-deconjugating enzymes from Arabidopsis
Abstract
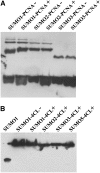
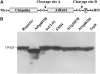
Posttranslational protein modification by the small ubiquitin-like modifier (SUMO) is a highly dynamic and reversible process. To analyze the substrate specificity of SUMO-conjugating and -deconjugating enzymes from Arabidopsis (Arabidopsis thaliana), we reconstituted its SUMOylation cascade in vitro and tested the capacity of this system to conjugate the Arabidopsis SUMO isoforms AtSUMO1, 2, and 3 to the model substrate ScPCNA from yeast (Saccharomyces cerevisiae). This protein contains two in vivo SUMOylated lysine residues, namely K127 and K164. Under in vitro conditions, the Arabidopsis SUMOylation system specifically conjugates all tested SUMO isoforms to lysine-127, but not to lysine-164, of ScPCNA. The SUMO isoforms AtSUMO1 and AtSUMO2, but not AtSUMO3, were found to form polymeric chains on ScPCNA due to a self-SUMOylation process. In a complementary approach, we analyzed both the SUMO isopeptidase activity and the pre-SUMO-processing capacity of the putative Arabidopsis SUMO proteases At1g60220, At1g10570, and At5g60190 using the known SUMO isopeptidases ScULP1, XopD, and ESD4 (At4g15880) as reference enzymes. Interestingly, At5g60190 exhibits no SUMO protease activity but processes the pre-form of Arabidopsis Rub1. The other five enzymes represent SUMO isopeptidases that show different substrate preferences. All these enzymes cleave AtSUMO1 and AtSUMO2 conjugates of ScPCNA, whereas only the putative bacterial virulence factor XopD is able to release AtSUMO3. In addition, all five enzymes cleave pre-AtSUMO1 and pre-AtSUMO2 peptides, but none of the proteins efficiently produce mature AtSUMO3 or AtSUMO5 molecules from their precursors.
Figures






Similar articles
-
Reconstitution of Arabidopsis thaliana SUMO pathways in E. coli: functional evaluation of SUMO machinery proteins and mapping of SUMOylation sites by mass spectrometry.Plant Cell Physiol. 2009 Jun;50(6):1049-61. doi: 10.1093/pcp/pcp056. Epub 2009 Apr 17. Plant Cell Physiol. 2009. PMID: 19376783
-
Small ubiquitin-like modifier modulates abscisic acid signaling in Arabidopsis.Plant Cell. 2003 Jun;15(6):1347-59. doi: 10.1105/tpc.009902. Plant Cell. 2003. PMID: 12782728 Free PMC article.
-
Arabidopsis thaliana proliferating cell nuclear antigen has several potential sumoylation sites.J Exp Bot. 2012 May;63(8):2971-83. doi: 10.1093/jxb/ers002. Epub 2012 Feb 13. J Exp Bot. 2012. PMID: 22330895
-
SUMO-specific proteases: a twist in the tail.Trends Cell Biol. 2007 Aug;17(8):370-6. doi: 10.1016/j.tcb.2007.08.002. Epub 2007 Sep 4. Trends Cell Biol. 2007. PMID: 17768054 Review.
-
Diversity of the SUMOylation machinery in plants.Biochem Soc Trans. 2010 Feb;38(Pt 1):60-4. doi: 10.1042/BST0380060. Biochem Soc Trans. 2010. PMID: 20074036 Review.
Cited by
-
The conjugation of SUMO to the transcription factor MYC2 functions in blue light-mediated seedling development in Arabidopsis.Plant Cell. 2022 Jul 30;34(8):2892-2906. doi: 10.1093/plcell/koac142. Plant Cell. 2022. PMID: 35567527 Free PMC article.
-
Arabidopsis SUMO E3 ligase SIZ1 is involved in excess copper tolerance.Plant Physiol. 2011 Aug;156(4):2225-34. doi: 10.1104/pp.111.178996. Epub 2011 Jun 1. Plant Physiol. 2011. PMID: 21632972 Free PMC article.
-
Small ubiquitin-like modifier proteases OVERLY TOLERANT TO SALT1 and -2 regulate salt stress responses in Arabidopsis.Plant Cell. 2008 Oct;20(10):2894-908. doi: 10.1105/tpc.108.058669. Epub 2008 Oct 10. Plant Cell. 2008. PMID: 18849491 Free PMC article.
-
SUMOylome Profiling Reveals a Diverse Array of Nuclear Targets Modified by the SUMO Ligase SIZ1 during Heat Stress.Plant Cell. 2018 May;30(5):1077-1099. doi: 10.1105/tpc.17.00993. Epub 2018 Mar 27. Plant Cell. 2018. PMID: 29588388 Free PMC article.
-
The Ubiquitin Conjugating Enzyme: An Important Ubiquitin Transfer Platform in Ubiquitin-Proteasome System.Int J Mol Sci. 2020 Apr 21;21(8):2894. doi: 10.3390/ijms21082894. Int J Mol Sci. 2020. PMID: 32326224 Free PMC article. Review.
References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 - PubMed
-
- Bayer P, Arndt A, Metzger S, Mahajan R, Melchior F, Jaenicke R, Becker J (1998) Structure determination of the small ubiquitin-related modifier SUMO-1. J Mol Biol 280: 275–286 - PubMed
-
- Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD (2002) Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108: 345–356 - PubMed
-
- Bohren KM, Nadkarni V, Song JH, Gabbay KH, Owerbach D (2004) A M55V polymorphism in a novel SUMO gene (SUMO-4) differentially activates heat shock transcription factors and is associated with susceptibility to type I diabetes mellitus. J Biol Chem 279: 27233–27238 - PubMed
-
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

