Mass spectrometry-based metabolomics
- PMID: 16921475
- PMCID: PMC1904337
- DOI: 10.1002/mas.20108
Mass spectrometry-based metabolomics
Abstract
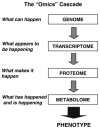
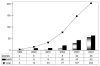
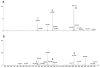
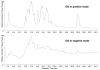
This review presents an overview of the dynamically developing field of mass spectrometry-based metabolomics. Metabolomics aims at the comprehensive and quantitative analysis of wide arrays of metabolites in biological samples. These numerous analytes have very diverse physico-chemical properties and occur at different abundance levels. Consequently, comprehensive metabolomics investigations are primarily a challenge for analytical chemistry and specifically mass spectrometry has vast potential as a tool for this type of investigation. Metabolomics require special approaches for sample preparation, separation, and mass spectrometric analysis. Current examples of those approaches are described in this review. It primarily focuses on metabolic fingerprinting, a technique that analyzes all detectable analytes in a given sample with subsequent classification of samples and identification of differentially expressed metabolites, which define the sample classes. To perform this complex task, data analysis tools, metabolite libraries, and databases are required. Therefore, recent advances in metabolomics bioinformatics are also discussed.
Copyright 2006 Wiley Periodicals, Inc.
Figures




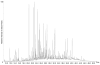
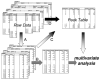
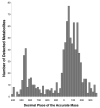
References
-
- Adahchour M, Brandt M, Baier HU, Vreuls RJ, Batenburg AM, Brinkman UA. Comprehensive two-dimensional gas chromatography coupled to a rapid-scanning quadrupole mass spectrometer: Principles and applications. J Chromatogr A. 2005;1067:245–254. - PubMed
-
- Aharoni A, Ric de Vos CH, Verhoeven HA, Maliepaard CA, Kruppa G, Bino R, Goodenowe DB. Nontargeted metabolome analysis by use of Fourier transform ion cyclotron mass spectrometry. Omics. 2002;6:217–234. - PubMed
-
- Allen J, Davey HM, Broadhurst D, Heald JK, Rowland JJ, Oliver SG, Kell DB. High-throughput classification of yeast mutants for functional genomics using metabolic footprinting. Nat Biotechnol. 2003;21:692–696. - PubMed
-
- Balazy M. Eicosanomics: Targeted lipidomics of eicosanoids in biological systems. Prostaglandins Other Lipid Mediat. 2004;73:173–180. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

