Rubredoxin:oxygen oxidoreductase enhances survival of Desulfovibrio vulgaris hildenborough under microaerophilic conditions
- PMID: 16923892
- PMCID: PMC1595363
- DOI: 10.1128/JB.00425-06
Rubredoxin:oxygen oxidoreductase enhances survival of Desulfovibrio vulgaris hildenborough under microaerophilic conditions
Abstract
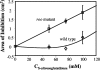
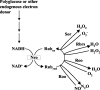
Genes for superoxide reductase (Sor), rubredoxin (Rub), and rubredoxin:oxygen oxidoreductase (Roo) are located in close proximity in the chromosome of Desulfovibrio vulgaris Hildenborough. Protein blots confirmed the absence of Roo from roo mutant and sor-rub-roo (srr) mutant cells and its presence in sor mutant and wild-type cells grown under anaerobic conditions. Oxygen reduction rates of the roo and srr mutants were 20 to 40% lower than those of the wild type and the sor mutant, indicating that Roo functions as an O2 reductase in vivo. Survival of single cells incubated for 5 days on agar plates under microaerophilic conditions (1% air) was 85% for the sor, 4% for the roo, and 0.7% for the srr mutant relative to that of the wild type (100%). The similar survival rates of sor mutant and wild-type cells suggest that O2 reduction by Roo prevents the formation of reactive oxygen species (ROS) under these conditions; i.e., the ROS-reducing enzyme Sor is only needed for survival when Roo is missing. In contrast, the sor mutant was inactivated much more rapidly than the roo mutant when liquid cultures were incubated in 100% air, indicating that O2 reduction by Roo and other terminal oxidases did not prevent ROS formation under these conditions. Competition of Sor and Roo for limited reduced Rub was suggested by the observation that the roo mutant survived better than the wild type under fully aerobic conditions. The roo mutant was more strongly inhibited than the wild type by the nitric oxide (NO)-generating compound S-nitrosoglutathione, indicating that Roo may also serve as an NO reductase in vivo.
Figures




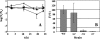

Similar articles
-
Function of oxygen resistance proteins in the anaerobic, sulfate-reducing bacterium Desulfovibrio vulgaris hildenborough.J Bacteriol. 2003 Jan;185(1):71-9. doi: 10.1128/JB.185.1.71-79.2003. J Bacteriol. 2003. PMID: 12486042 Free PMC article.
-
Contribution of rubredoxin:oxygen oxidoreductases and hybrid cluster proteins of Desulfovibrio vulgaris Hildenborough to survival under oxygen and nitrite stress.Environ Microbiol. 2012 Oct;14(10):2711-25. doi: 10.1111/j.1462-2920.2012.02859.x. Epub 2012 Sep 4. Environ Microbiol. 2012. PMID: 22947039
-
Desulfovibrio gigas flavodiiron protein affords protection against nitrosative stress in vivo.J Bacteriol. 2006 Apr;188(8):2745-51. doi: 10.1128/JB.188.8.2745-2751.2006. J Bacteriol. 2006. PMID: 16585735 Free PMC article.
-
Superoxide reductase: fact or fiction?J Biol Inorg Chem. 2002 Jun;7(6):647-52. doi: 10.1007/s00775-002-0359-x. Epub 2002 Apr 18. J Biol Inorg Chem. 2002. PMID: 12072972 Review.
-
The adaptive genome of Desulfovibrio vulgaris Hildenborough.FEMS Microbiol Lett. 2006 Jul;260(2):127-33. doi: 10.1111/j.1574-6968.2006.00261.x. FEMS Microbiol Lett. 2006. PMID: 16842335 Review.
Cited by
-
Cell-wide responses to low-oxygen exposure in Desulfovibrio vulgaris Hildenborough.J Bacteriol. 2007 Aug;189(16):5996-6010. doi: 10.1128/JB.00368-07. Epub 2007 Jun 1. J Bacteriol. 2007. PMID: 17545284 Free PMC article.
-
An Abundant and Diverse New Family of Electron Bifurcating Enzymes With a Non-canonical Catalytic Mechanism.Front Microbiol. 2022 Jul 8;13:946711. doi: 10.3389/fmicb.2022.946711. eCollection 2022. Front Microbiol. 2022. PMID: 35875533 Free PMC article.
-
The Selenoproteome as a Dynamic Response Mechanism to Oxidative Stress in Hydrogenotrophic Methanogenic Communities.Environ Sci Technol. 2024 Apr 16;58(15):6637-6646. doi: 10.1021/acs.est.3c07725. Epub 2024 Apr 5. Environ Sci Technol. 2024. PMID: 38580315 Free PMC article.
-
Insights into the nitric oxide reductase mechanism of flavodiiron proteins from a flavin-free enzyme.Biochemistry. 2010 Aug 24;49(33):7040-9. doi: 10.1021/bi100788y. Biochemistry. 2010. PMID: 20669924 Free PMC article.
-
Analysis of a ferric uptake regulator (Fur) mutant of Desulfovibrio vulgaris Hildenborough.Appl Environ Microbiol. 2007 Sep;73(17):5389-400. doi: 10.1128/AEM.00276-07. Epub 2007 Jul 13. Appl Environ Microbiol. 2007. PMID: 17630305 Free PMC article.
References
-
- Agostinho, M., S. Oliveira, M. Broco, M.-Y. Liu, J. LeGall, and C. Rodrigues-Pousada. 2000. Molecular cloning of the gene encoding flavoredoxin, a flavoprotein from Desulfovibrio gigas. Biochem. Biophys. Res. Commun. 272:653-656. - PubMed
-
- Broco, M., M. Rousset, S. Oliveira, and C. Rodrigues-Pousada. 2005. Deletion of flavoredoxin gene in Desulfovibrio gigas reveals its participation in thiosulfate reduction. FEBS Lett. 579:4803-4807. - PubMed
-
- Chen, L., M.-Y Liu, J. LeGall, P. Fareleira, H. Santos, and A. V. Xavier. 1993. Rubredoxin oxidase, a new flavo-hemo protein, is the site of oxygen reduction to water by the “strict anaerobe” Desulfovibrio gigas. Biochem. Biophys. Res. Commun. 193:100-105. - PubMed
-
- Coulter, E. D., and D. M. Kurtz, Jr. 2001. A role for rubredoxin in oxidative stress protection in Desulfovibrio vulgaris: catalytic electron transfer to rubrerythrin and two-iron superoxide reductase. Arch. Biochem. Biophys. 394:76-86. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

