Regulation of ppk expression and in vivo function of Ppk in Streptomyces lividans TK24
- PMID: 16923894
- PMCID: PMC1595360
- DOI: 10.1128/JB.00202-06
Regulation of ppk expression and in vivo function of Ppk in Streptomyces lividans TK24
Abstract
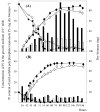
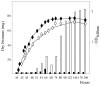
The ppk gene of Streptomyces lividans encodes an enzyme catalyzing, in vitro, the reversible polymerization of the gamma phosphate of ATP into polyphosphate and was previously shown to play a negative role in the control of antibiotic biosynthesis (H. Chouayekh and M. J. Virolle, Mol. Microbiol. 43:919-930, 2002). In the present work, some regulatory features of the expression of ppk were established and the polyphosphate content of S. lividans TK24 and the ppk mutant was determined. In Pi sufficiency, the expression of ppk was shown to be low but detectable. DNA gel shift experiments suggested that ppk expression might be controlled by a repressor using ATP as a corepressor. Under these conditions, short acid-soluble polyphosphates accumulated upon entry into the stationary phase in the wild-type strain but not in the ppk mutant strain. The expression of ppk under Pi-limiting conditions was shown to be much higher than that under Pi-sufficient conditions and was under positive control of the two-component system PhoR/PhoP. Under these conditions, the polyphosphate content of the cell was low and polyphosphates were reproducibly found to be longer and more abundant in the ppk mutant strain than in the wild-type strain, suggesting that Ppk might act as a nucleoside diphosphate kinase. In light of our results, a novel view of the role of this enzyme in the regulation of antibiotic biosynthesis in S. lividans TK24 is proposed.
Figures




References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1995. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
-
- Avignone Rossa, C., J. White, A. Kuiper, P. W. Postma, M. Bibb, and M. J. Teixeira de Mattos. 2002. Carbon flux distribution in antibiotic-producing chemostat cultures of Streptomyces lividans. Metab. Eng. 4:138-150. - PubMed
-
- Bibb, M. J. 2005. Regulation of secondary metabolism in streptomycetes. Curr. Opin. Microbiol. 8:208-215. - PubMed
-
- Blondelet-Rouault, M. H., J. Weiser, A. Lebrihi, P. Branny, and J. L. Pernodet. 1997. Antibiotic resistance gene cassettes derived from the omega interposon for use in E. coli and Streptomyces. Gene 190:315-317. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

