The highly efficient translation initiation region from the Escherichia coli rpsA gene lacks a shine-dalgarno element
- PMID: 16923895
- PMCID: PMC1595398
- DOI: 10.1128/JB.00591-06
The highly efficient translation initiation region from the Escherichia coli rpsA gene lacks a shine-dalgarno element
Abstract
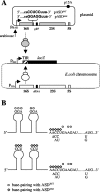
The translational initiation region (TIR) of the Escherichia coli rpsA gene, which encodes ribosomal protein S1, shows a number of unusual features. It extends far upstream (to position -91) of the initiator AUG, it lacks a canonical Shine-Dalgarno sequence (SD) element, and it can fold into three successive hairpins (I, II, and III) that are essential for high translational activity. Two conserved GGA trinucleotides, present in the loops of hairpins I and II, have been proposed to form a discontinuous SD. Here, we have tested this hypothesis with the "specialized ribosome" approach. Depending upon the constructs used, translation initiation was decreased three- to sevenfold upon changing the conserved GGA to CCU. However, although chemical probing showed that the mutated trinucleotides were accessible, no restoration was observed when the ribosome anti-SD was symmetrically changed from CCUCC to GGAGG. When the same change was introduced in the SD from a conventional TIR as a control, activity was stimulated. This result suggests that the GGA trinucleotides do not form a discontinuous SD. Others hypotheses that may account for their role are discussed. Curiously, we also find that, when expressed at moderate level (30 to 40% of total ribosomes), specialized ribosomes are only twofold disadvantaged over normal ribosomes for the translation of bulk cellular mRNAs. These findings suggest that, under these conditions, the SD-anti-SD interaction plays a significant but not essential role for the synthesis of bulk cellular proteins.
Figures




References
-
- Bélanger, F., M. Léger, A. A. Saraiya, P. R. Cunningham, and L. Brakier-Gingras. 2002. Functional studies of the 900 tetraloop capping helix 27 of 16S ribosomal RNA. J. Mol. Biol. 320:979-989. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

