Genetic analysis of the requirement for flp-2, tadV, and rcpB in Actinobacillus actinomycetemcomitans biofilm formation
- PMID: 16923904
- PMCID: PMC1595400
- DOI: 10.1128/JB.00496-06
Genetic analysis of the requirement for flp-2, tadV, and rcpB in Actinobacillus actinomycetemcomitans biofilm formation
Abstract
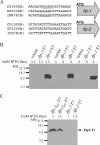
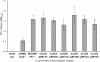
The tad locus of Actinobacillus actinomycetemcomitans encodes a molecular transport system required for tenacious, nonspecific adherence to surfaces and formation of extremely strong biofilms. This locus is dedicated to the biogenesis of Flp pili, which are required for colonization and virulence. We have previously shown that 11 of the 14 tad locus genes are required for adherence and Flp pilus production. Here, we present genetic and phylogenetic analyses of flp-2, tadV, and rcpB genes in biofilm formation. We show that tadV, predicted to encode prepilin peptidase, is required for adherence. In contrast, targeted insertional inactivation of flp-2, a gene closely related to the prepillin gene flp-1, did not abrogate biofilm formation. Expression studies did not detect Flp2-T7 protein under standard laboratory conditions. We present phylogenetic data showing that there is no significant evidence for natural selection in the available flp-2 sequences from A. actinomycetemcomitans, suggesting that flp-2 does not play a significant role in the biology of this organism. Mutants with insertions at the 3' end of rcpB formed biofilms equivalent to wild-type A. actinomycetemcomitans. Surprisingly, 5' end chromosomal insertion mutants in rcpB were obtained only when a wild-type copy of the rcpB gene was provided in trans or when the Tad secretion system was inactivated. Together, our results strongly suggest that A. actinomycetemcomitans rcpB is essential in the context of a functional tad locus. These data show three different phenotypes for the three genes.
Figures






References
-
- Das, M., A. D. Badley, F. R. Cockerill, J. M. Steckelberg, and W. R. Wilson. 1997. Infective endocarditis caused by HACEK microorganisms. Annu. Rev. Med. 48:25-33. - PubMed
-
- Eriksson, T. 1997. Autodecay, 2.9.9 ed. Hypercard stack distributed by the author. Botaniska Institutionen, Stockholm University, Stockholm, Sweden.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

