Bacillus subtilis aconitase is required for efficient late-sporulation gene expression
- PMID: 16923907
- PMCID: PMC1595401
- DOI: 10.1128/JB.00249-06
Bacillus subtilis aconitase is required for efficient late-sporulation gene expression
Erratum in
- J Bacteriol. 2007 Jul;189(14):5403
Abstract
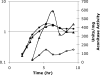
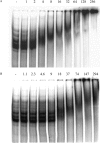
Bacillus subtilis aconitase, encoded by the citB gene, is homologous to the bifunctional eukaryotic protein IRP-1 (iron regulatory protein 1). Like IRP-1, B. subtilis aconitase is both an enzyme and an RNA binding protein. In an attempt to separate the two activities of aconitase, the C-terminal region of the B. subtilis citB gene product was mutagenized. The resulting strain had high catalytic activity but was defective in sporulation. The defect was at a late stage of sporulation, specifically affecting expression of sigmaK-dependent genes, many of which are important for spore coat assembly and require transcriptional activation by GerE. Accumulation of gerE mRNA and GerE protein was delayed in the aconitase mutant strain. Pure B. subtilis aconitase bound to the 3' untranslated region of gerE mRNA in in vitro gel mobility shift assays, strongly suggesting that aconitase RNA binding activity may stabilize gerE mRNA in order to allow efficient GerE synthesis and proper timing of spore coat assembly.
Figures







References
-
- Addess, K. J., J. P. Basilion, R. D. Klausner, T. A. Rouault, and A. Pardi. 1997. Structure and dynamics of the iron responsive element RNA: implications for binding of the RNA by iron regulatory binding proteins. J. Mol. Biol. 274:72-83. - PubMed
-
- Bulteau, A. L., H. A. O'Neill, M. C. Kennedy, M. Ikeda-Saito, G. Isaya, and L. I. Szweda. 2004. Frataxin acts as an iron chaperone protein to modulate mitochondrial aconitase activity. Science 305:242-245. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

