Genetic evidence for an interaction of the UbiG O-methyltransferase with UbiX in Escherichia coli coenzyme Q biosynthesis
- PMID: 16923914
- PMCID: PMC1595381
- DOI: 10.1128/JB.00668-06
Genetic evidence for an interaction of the UbiG O-methyltransferase with UbiX in Escherichia coli coenzyme Q biosynthesis
Abstract
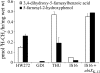
IS16 is a thiol-sensitive, Q-deficient mutant strain of Escherichia coli. Here, we show that IS16 harbors a mutation in the ubiG gene encoding a methyltransferase required for two O-methylation steps of Q biosynthesis. Complementation of IS16 with either ubiG or ubiX(K-12) reverses this phenotype, suggesting that UbiX may interact with UbiG.
Figures





Similar articles
-
The role of UbiX in Escherichia coli coenzyme Q biosynthesis.Arch Biochem Biophys. 2007 Nov 15;467(2):144-53. doi: 10.1016/j.abb.2007.08.009. Epub 2007 Aug 23. Arch Biochem Biophys. 2007. PMID: 17889824 Free PMC article.
-
Complementation of coq3 mutant yeast by mitochondrial targeting of the Escherichia coli UbiG polypeptide: evidence that UbiG catalyzes both O-methylation steps in ubiquinone biosynthesis.Biochemistry. 1996 Jul 30;35(30):9797-806. doi: 10.1021/bi9602932. Biochemistry. 1996. PMID: 8703953
-
Restoring de novo coenzyme Q biosynthesis in Caenorhabditis elegans coq-3 mutants yields profound rescue compared to exogenous coenzyme Q supplementation.Gene. 2012 Sep 10;506(1):106-16. doi: 10.1016/j.gene.2012.06.023. Epub 2012 Jun 23. Gene. 2012. PMID: 22735617 Free PMC article.
-
Biosynthesis and physiology of coenzyme Q in bacteria.Biochim Biophys Acta. 2014 Jul;1837(7):1004-11. doi: 10.1016/j.bbabio.2014.01.015. Epub 2014 Jan 28. Biochim Biophys Acta. 2014. PMID: 24480387 Review.
-
The UbiX-UbiD system: The biosynthesis and use of prenylated flavin (prFMN).Arch Biochem Biophys. 2017 Oct 15;632:209-221. doi: 10.1016/j.abb.2017.07.014. Epub 2017 Jul 25. Arch Biochem Biophys. 2017. PMID: 28754323 Review.
Cited by
-
Chorismate pyruvate-lyase and 4-hydroxy-3-solanesylbenzoate decarboxylase are required for plastoquinone biosynthesis in the cyanobacterium Synechocystis sp. PCC6803.J Biol Chem. 2014 Jan 31;289(5):2675-86. doi: 10.1074/jbc.M113.511709. Epub 2013 Dec 11. J Biol Chem. 2014. PMID: 24337576 Free PMC article.
-
Biosynthesis of Menaquinone (Vitamin K2) and Ubiquinone (Coenzyme Q).EcoSal Plus. 2009 Aug;3(2):10.1128/ecosalplus.3.6.3.3. doi: 10.1128/ecosalplus.3.6.3.3. EcoSal Plus. 2009. PMID: 26443765 Free PMC article.
-
The role of UbiX in Escherichia coli coenzyme Q biosynthesis.Arch Biochem Biophys. 2007 Nov 15;467(2):144-53. doi: 10.1016/j.abb.2007.08.009. Epub 2007 Aug 23. Arch Biochem Biophys. 2007. PMID: 17889824 Free PMC article.
-
Crystallization and preliminary crystallographic studies of UbiG, an O-methyltransferase from Escherichia coli.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011 Jun 1;67(Pt 6):727-9. doi: 10.1107/S1744309111014278. Epub 2011 May 26. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011. PMID: 21636923 Free PMC article.
References
-
- Baba, S. W., G. I. Belogrudov, J. C. Lee, P. T. Lee, J. Strahan, J. N. Shepherd, and C. F. Clarke. 2004. Yeast Coq5 C-methyltransferase is required for stability of other polypeptides involved in coenzyme Q biosynthesis. J. Biol. Chem. 279:10052-10059. - PubMed
-
- Bader, M., W. Muse, D. P. Ballou, C. Gassner, and J. C. Bardwell. 1999. Oxidative protein folding is driven by the electron transport system. Cell 98:217-227. - PubMed
-
- Belogrudov, G. I., P. T. Lee, T. Jonassen, A. Y. Hsu, P. Gin, and C. F. Clarke. 2001. Yeast COQ4 encodes a mitochondrial protein required for coenzyme Q. synthesis. Arch. Biochem. Biophys. 392:48-58. - PubMed
-
- Brandt, U., and B. Trumpower. 1994. The protonmotive Q cycle in mitochondria and bacteria. Crit. Rev. Biochem. Mol. Biol. 29:165-197. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

