Role of topoisomerase IIbeta in the expression of developmentally regulated genes
- PMID: 16923961
- PMCID: PMC1636731
- DOI: 10.1128/MCB.00617-06
Role of topoisomerase IIbeta in the expression of developmentally regulated genes
Abstract
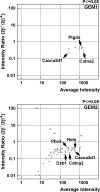
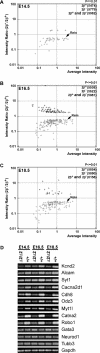
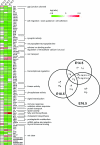
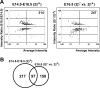
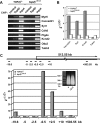
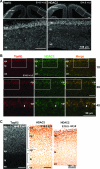
Mice lacking topoisomerase IIbeta (TopIIbeta) are known to exhibit a perinatal death phenotype. In the current study, transcription profiles of the brains of wild-type and top2beta knockout mouse embryos were generated. Surprisingly, only a small number (1 to 4%) of genes were affected in top2beta knockout embryos. However, the expression of nearly 30% of developmentally regulated genes was either up- or down-regulated. By contrast, the expression of genes encoding general cell growth functions and early differentiation markers was not affected, suggesting that TopIIbeta is not required for early differentiation programming but is specifically required for the expression of developmentally regulated genes at later stages of differentiation. Consistent with this notion, immunohistochemical analysis of brain sections showed that TopIIbeta and histone deacetylase 2, a known TopIIbeta-interacting protein, were preferentially expressed in neurons which are in their later stages of differentiation. Chromatin immunoprecipitation analysis of the developing brains revealed TopIIbeta binding to the 5' region of a number of TopIIbeta-sensitive genes. Further studies of a TopIIbeta-sensitive gene, Kcnd2, revealed the presence of TopIIbeta in the transcription unit with major binding near the promoter region. Together, these results support a role of TopIIbeta in activation/repression of developmentally regulated genes at late stages of neuronal differentiation.
Figures

References
-
- Allen, M. P., M. Xu, C. Zeng, S. A. Tobet, and M. E. Wierman. 2000. Myocyte enhancer factors-2B and -2C are required for adhesion related kinase repression of neuronal gonadotropin releasing hormone gene expression. J. Biol. Chem. 275:39662-39670. - PubMed
-
- Austin, C. A., and K. L. Marsh. 1998. Eukaryotic DNA topoisomerase II beta. Bioessays 20:215-226. - PubMed
-
- Austin, C. A., J. H. Sng, S. Patel, and L. M. Fisher. 1993. Novel HeLa topoisomerase II is the II beta isoform: complete coding sequence and homology with other type II topoisomerases. Biochim. Biophys. Acta 1172:283-291. - PubMed
-
- Bhat, M. A., A. V. Philp, D. M. Glover, and H. J. Bellen. 1996. Chromatid segregation at anaphase requires the barren product, a novel chromosome-associated protein that interacts with topoisomerase II. Cell 87:1103-1114. - PubMed
-
- Boulikas, T. 1995. Chromatin domains and prediction of MAR sequences. Int. Rev. Cytol. 162A:279-388. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
