Towards synthesis of a minimal cell
- PMID: 16924266
- PMCID: PMC1681520
- DOI: 10.1038/msb4100090
Towards synthesis of a minimal cell
Abstract
Construction of a chemical system capable of replication and evolution, fed only by small molecule nutrients, is now conceivable. This could be achieved by stepwise integration of decades of work on the reconstitution of DNA, RNA and protein syntheses from pure components. Such a minimal cell project would initially define the components sufficient for each subsystem, allow detailed kinetic analyses and lead to improved in vitro methods for synthesis of biopolymers, therapeutics and biosensors. Completion would yield a functionally and structurally understood self-replicating biosystem. Safety concerns for synthetic life will be alleviated by extreme dependence on elaborate laboratory reagents and conditions for viability. Our proposed minimal genome is 113 kbp long and contains 151 genes. We detail building blocks already in place and major hurdles to overcome for completion.
Figures
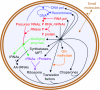
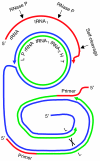
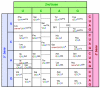
References
-
- Andachi Y, Yamao F, Muto A, Osawa S (1989) Codon recognition patterns as deduced from sequences of the complete set of transfer RNA species in Mycoplasma capricolum. Resemblance to mitochondria. J Mol Biol 209: 37–54 - PubMed
-
- Bjork GR (1995) Biosynthesis and function of modified nucleosides. In Dieter Söll, Uttam RajBhandary (eds), tRNA: Structure, Biosynthesis, and Function, pp 165–205. Washington, DC: ASM Press
-
- Cho MK, Magnus D, Caplan AL, McGee D (1999) Policy forum: genetics. Ethical considerations in synthesizing a minimal genome. Science 286: 2087–2090 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

