Mechanisms of RecQ helicases in pathways of DNA metabolism and maintenance of genomic stability
- PMID: 16925525
- PMCID: PMC1559444
- DOI: 10.1042/BJ20060450
Mechanisms of RecQ helicases in pathways of DNA metabolism and maintenance of genomic stability
Abstract
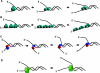
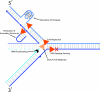
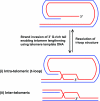
Helicases are molecular motor proteins that couple the hydrolysis of NTP to nucleic acid unwinding. The growing number of DNA helicases implicated in human disease suggests that their vital specialized roles in cellular pathways are important for the maintenance of genome stability. In particular, mutations in genes of the RecQ family of DNA helicases result in chromosomal instability diseases of premature aging and/or cancer predisposition. We will discuss the mechanisms of RecQ helicases in pathways of DNA metabolism. A review of RecQ helicases from bacteria to human reveals their importance in genomic stability by their participation with other proteins to resolve DNA replication and recombination intermediates. In the light of their known catalytic activities and protein interactions, proposed models for RecQ function will be summarized with an emphasis on how this distinct class of enzymes functions in chromosomal stability maintenance and prevention of human disease and cancer.
Figures


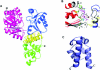


References
-
- Harrigan J. A., Bohr V. A. Human diseases deficient in RecQ helicases. Biochimie. 2003;85:1185–1193. - PubMed
-
- Siitonen H. A., Kopra O., Kaariainen H., Haravuori H., Winter R. M., Säämänen A. M., Peltonen L., Kestila M. Molecular defect of RAPADILINO syndrome expands the phenotype spectrum of RECQL diseases. Hum. Mol. Genet. 2003;12:2837–2844. - PubMed
-
- Van Maldergem L., Siitonen H. A., Jalkh N., Chouery N., De Roy M., Delague V., Muenke M., Jabs E. W., Cai J., Wang L. L., et al. Revisiting the craniosynostosis-radial ray hypoplasia association: Baller–Gerold syndrome caused by mutations in the RECQL4 gene. J. Med. Genet. 2006;43:148–152. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

