Identification by functional proteomics of a deubiquitinating/deNeddylating enzyme in Plasmodium falciparum
- PMID: 16925553
- PMCID: PMC7168409
- DOI: 10.1111/j.1365-2958.2006.05307.x
Identification by functional proteomics of a deubiquitinating/deNeddylating enzyme in Plasmodium falciparum
Abstract
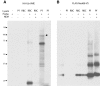
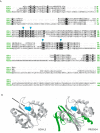
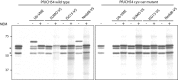
Ubiquitination is a post-translational modification implicated in a variety of cellular functions, including transcriptional regulation, protein degradation and membrane protein trafficking. Ubiquitin and the enzymes that act on it, although conserved and essential in eukaryotes, have not been well studied in parasites, despite sequencing of several parasite genomes. Several putative ubiquitin hydrolases have been identified in Plasmodium falciparum based on sequence homology alone, with no evidence of expression or function. Here we identify the first deubiquitinating enzyme in P. falciparum, PfUCH54, by its activity. We show that PfUCH54 also has deNeddylating activity, as assayed by a mammalian Nedd8-based probe. This activity is absent from mammalian homologues of PfUCH54. Given the importance of parasitic membrane protein trafficking as well as protein degradation in the virulence of this parasite, this family of enzymes may represent a target for pharmacological intervention with this disease.
Figures

References
-
- Binder, E.M. , and Kim, K. (2004) Location, location, location: trafficking and function of secreted proteases of Toxoplasma and Plasmodium . Traffic 5: 914–924. - PubMed
-
- Borodovsky, A. , Ovaa, H. , Kolli, N. , Gan‐Erdene, T. , Wilkinson, K.D. , Ploegh, H.L. , and Kessler, B.M. (2002) Chemistry‐based functional proteomics reveals novel members of the deubiquitinating enzyme. Chem Biol 9: 1149–1159. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

