Assessment of whole genome amplification-induced bias through high-throughput, massively parallel whole genome sequencing
- PMID: 16928277
- PMCID: PMC1560136
- DOI: 10.1186/1471-2164-7-216
Assessment of whole genome amplification-induced bias through high-throughput, massively parallel whole genome sequencing
Abstract
Background: Whole genome amplification is an increasingly common technique through which minute amounts of DNA can be multiplied to generate quantities suitable for genetic testing and analysis. Questions of amplification-induced error and template bias generated by these methods have previously been addressed through either small scale (SNPs) or large scale (CGH array, FISH) methodologies. Here we utilized whole genome sequencing to assess amplification-induced bias in both coding and non-coding regions of two bacterial genomes. Halobacterium species NRC-1 DNA and Campylobacter jejuni were amplified by several common, commercially available protocols: multiple displacement amplification, primer extension pre-amplification and degenerate oligonucleotide primed PCR. The amplification-induced bias of each method was assessed by sequencing both genomes in their entirety using the 454 Sequencing System technology and comparing the results with those obtained from unamplified controls.
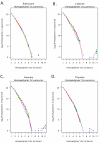
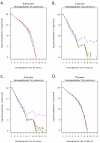
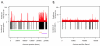
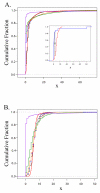
Results: All amplification methodologies induced statistically significant bias relative to the unamplified control. For the Halobacterium species NRC-1 genome, assessed at 100 base resolution, the D-statistics from GenomiPhi-amplified material were 119 times greater than those from unamplified material, 164.0 times greater for Repli-G, 165.0 times greater for PEP-PCR and 252.0 times greater than the unamplified controls for DOP-PCR. For Campylobacter jejuni, also analyzed at 100 base resolution, the D-statistics from GenomiPhi-amplified material were 15 times greater than those from unamplified material, 19.8 times greater for Repli-G, 61.8 times greater for PEP-PCR and 220.5 times greater than the unamplified controls for DOP-PCR.
Conclusion: Of the amplification methodologies examined in this paper, the multiple displacement amplification products generated the least bias, and produced significantly higher yields of amplified DNA.
Figures


Similar articles
-
Characterization of whole genome amplified (WGA) DNA for use in genotyping assay development.BMC Genomics. 2012 Jun 1;13:217. doi: 10.1186/1471-2164-13-217. BMC Genomics. 2012. PMID: 22655855 Free PMC article.
-
Assessment of REPLI-g Multiple Displacement Whole Genome Amplification (WGA) Techniques for Metagenomic Applications.J Biomol Tech. 2017 Apr;28(1):46-55. doi: 10.7171/jbt.17-2801-008. Epub 2017 Mar 21. J Biomol Tech. 2017. PMID: 28344519 Free PMC article.
-
Evaluation of Phi29-based whole-genome amplification for microarray-based comparative genomic hybridisation.Lab Invest. 2007 Jan;87(1):75-83. doi: 10.1038/labinvest.3700495. Lab Invest. 2007. PMID: 17170740
-
Bias in Whole Genome Amplification: Causes and Considerations.Methods Mol Biol. 2015;1347:15-41. doi: 10.1007/978-1-4939-2990-0_2. Methods Mol Biol. 2015. PMID: 26374307 Review.
-
[Comparison of different single cell whole genome amplification methods and MALBAC applications in assisted reproduction].Yi Chuan. 2018 Aug 16;40(8):620-631. doi: 10.16288/j.yczz.18-091. Yi Chuan. 2018. PMID: 30117418 Review. Chinese.
Cited by
-
A common founding clone with TP53 and PTEN mutations gives rise to a concurrent germ cell tumor and acute megakaryoblastic leukemia.Cold Spring Harb Mol Case Stud. 2016 Jan;2(1):a000687. doi: 10.1101/mcs.a000687. Cold Spring Harb Mol Case Stud. 2016. PMID: 27148581 Free PMC article.
-
The Production of Recombinant African Swine Fever Virus Lv17/WB/Rie1 Strains and Their In Vitro and In Vivo Characterizations.Vaccines (Basel). 2023 Dec 17;11(12):1860. doi: 10.3390/vaccines11121860. Vaccines (Basel). 2023. PMID: 38140263 Free PMC article.
-
Characterization of whole genome amplified (WGA) DNA for use in genotyping assay development.BMC Genomics. 2012 Jun 1;13:217. doi: 10.1186/1471-2164-13-217. BMC Genomics. 2012. PMID: 22655855 Free PMC article.
-
Metagenomics and future perspectives in virus discovery.Curr Opin Virol. 2012 Feb;2(1):63-77. doi: 10.1016/j.coviro.2011.12.004. Epub 2012 Jan 20. Curr Opin Virol. 2012. PMID: 22440968 Free PMC article. Review.
-
Chromosome arm-specific BAC end sequences permit comparative analysis of homoeologous chromosomes and genomes of polyploid wheat.BMC Plant Biol. 2012 May 4;12:64. doi: 10.1186/1471-2229-12-64. BMC Plant Biol. 2012. PMID: 22559868 Free PMC article.
References
-
- Andries K, Verhasselt P, Guillemont J, Gohlmann HW, Neefs JM, Winkler H, Van Gestel J, Timmerman P, Zhu M, Lee E, Williams P, de Chaffoy D, Huitric E, Hoffner S, Cambau E, Truffot-Pernot C, Lounis N, Jarlier V. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science. 2005;307:223–227. doi: 10.1126/science.1106753. - DOI - PubMed
-
- Zheng S, Ma X, Buffler PA, Smith MT, Wiencke JK. Whole genome amplification increases the efficiency and validity of buccal cell genotyping in pediatric populations. Cancer Epidemiol Biomarkers Prev. 2001;10:697–700. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

