Porcine endogenous retrovirus integration sites in the human genome: features in common with those of murine leukemia virus
- PMID: 16928752
- PMCID: PMC1642138
- DOI: 10.1128/JVI.00904-06
Porcine endogenous retrovirus integration sites in the human genome: features in common with those of murine leukemia virus
Abstract
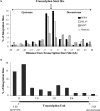
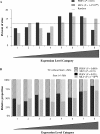
Porcine endogenous retroviruses (PERV) are a major concern when porcine tissues and organs are used for xenotransplantation. PERV has been shown to infect human cells in vitro, highlighting a potential zoonotic risk. No pathology is associated with PERV in its natural host, but the pathogenic potential might differ in the case of cross-species transmission and can only be inferred from knowledge of related gammaretroviruses. We therefore investigated the integration features of the PERV DNA in the human genome in vitro in order to further characterize the risk associated with PERV transmission. In this study, we characterized 189 PERV integration site sequences from human HEK-293 cells. Data showed that PERV integration was strongly enhanced at transcriptional start sites and CpG islands and that the frequencies of integration events increased with the expression levels of the genes, except for the genes with the highest levels of expression, which were disfavored for integration. Finally, we extracted genomic sequences directly flanking the integration sites and found an original 8-base statistical palindromic consensus sequence [TG(int)GTACCAGC]. All these results show similarities between PERV and murine leukemia virus integration site selection, suggesting that gammaretroviruses have a common pattern of integration and that the mechanisms of target site selection within a retrovirus genus might be similar.
Figures


References
-
- Bushman, F., M. Lewinski, A. Ciuffi, S. Barr, J. Leipzig, S. Hannenhalli, and C. Hoffmann. 2005. Genome-wide analysis of retroviral DNA integration. Nat. Rev. Microbiol. 3:848-858. - PubMed
-
- Bushman, F. D. 2003. Targeting survival: integration site selection by retroviruses and LTR-retrotransposons. Cell 115:135-138. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

