Viral and cellular determinants of the hepatitis C virus envelope-heparan sulfate interaction
- PMID: 16928753
- PMCID: PMC1641783
- DOI: 10.1128/JVI.00941-06
Viral and cellular determinants of the hepatitis C virus envelope-heparan sulfate interaction
Abstract
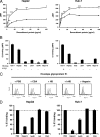
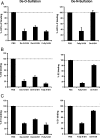
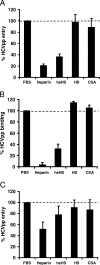
Cellular binding and entry of hepatitis C virus (HCV) are the first steps of viral infection and represent a major target for antiviral antibodies and novel therapeutic strategies. We have recently demonstrated that heparan sulfate (HS) plays a key role in the binding of HCV envelope glycoprotein E2 to target cells (Barth et al., J. Biol. Chem. 278:41003-41012, 2003). In this study, we characterized the HCV-HS interaction and analyzed its inhibition by antiviral host immune responses. Using recombinant envelope glycoproteins, virus-like particles, and HCV pseudoparticles as model systems for the early steps of viral infection, we mapped viral and cellular determinants of HCV-HS interaction. HCV-HS binding required a specific HS structure that included N-sulfo groups and a minimum of 10 to 14 saccharide subunits. HCV envelope binding to HS was mediated by four viral epitopes overlapping the E2 hypervariable region 1 and E2-CD81 binding domains. In functional studies using HCV pseudoparticles, we demonstrate that HCV binding and entry are specifically inhibited by highly sulfated HS. Finally, HCV-HS binding was markedly inhibited by antiviral antibodies derived from HCV-infected individuals. In conclusion, our results demonstrate that binding of the viral envelope to a specific HS configuration represents an important step for the initiation of viral infection and is a target of antiviral host immune responses in vivo. Mapping of viral and cellular determinants of HCV-HS interaction sets the stage for the development of novel HS-based antiviral strategies targeting viral attachment and entry.
Figures
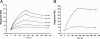



References
-
- Bain, C., A. Fatmi, F. Zoulim, J. P. Zarski, C. Trepo, and G. Inchauspe. 2001. Impaired allostimulatory function of dendritic cells in chronic hepatitis C infection. Gastroenterology 120:512-524. - PubMed
-
- Barth, H., C. Schafer, M. I. Adah, F. Zhang, R. J. Linhardt, H. Toyoda, A. Kinoshita-Toyoda, T. Toida, T. H. Van Kuppevelt, E. Depla, F. Von Weizsacker, H. E. Blum, and T. F. Baumert. 2003. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J. Biol. Chem. 278:41003-41012. - PubMed
-
- Bartosch, B., A. Vitelli, C. Granier, C. Goujon, J. Dubuisson, S. Pascale, E. Scarselli, R. Cortese, A. Nicosia, and F. L. Cosset. 2003. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J. Biol. Chem. 278:41624-41630. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

