Integrated molecular signature of disease: analysis of influenza virus-infected macaques through functional genomics and proteomics
- PMID: 16928763
- PMCID: PMC1641753
- DOI: 10.1128/JVI.00851-06
Integrated molecular signature of disease: analysis of influenza virus-infected macaques through functional genomics and proteomics
Abstract
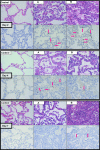
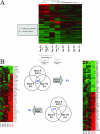
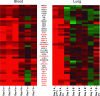
Recent outbreaks of avian influenza in humans have stressed the need for an improved nonhuman primate model of influenza pathogenesis. In order to further develop a macaque model, we expanded our previous in vivo genomics experiments with influenza virus-infected macaques by focusing on the innate immune response at day 2 postinoculation and on gene expression in affected lung tissue with viral genetic material present. Finally, we sought to identify signature genes for early infection in whole blood. For these purposes, we infected six pigtailed macaques (Macaca nemestrina) with reconstructed influenza A/Texas/36/91 virus and three control animals with a sham inoculate. We sacrificed one control and two experimental animals at days 2, 4, and 7 postinfection. Lung tissue was harvested for pathology, gene expression profiling, and proteomics. Blood was collected for genomics every other day from each animal until the experimental endpoint. Gross and microscopic pathology, immunohistochemistry, viral gene expression by arrays, and/or quantitative real-time reverse transcription-PCR confirmed successful yet mild infections in all experimental animals. Genomic experiments were performed using macaque-specific oligonucleotide arrays, and high-throughput proteomics revealed the host response to infection at the mRNA and protein levels. Our data showed dramatic differences in gene expression within regions in influenza virus-induced lesions based on the presence or absence of viral mRNA. We also identified genes tightly coregulated in peripheral white blood cells and in lung tissue at day 2 postinoculation. This latter finding opens the possibility of using gene expression arrays on whole blood to detect infection after exposure but prior to onset of symptoms or shedding.
Figures


References
-
- Aggarwal, S., S. Gollapudi, and S. Gupta. 1999. Increased TNF-alpha-induced apoptosis in lymphocytes from aged humans: changes in TNF-alpha receptor expression and activation of caspases. J. Immunol. 162:2154-2161. - PubMed
-
- Barnes, B., B. Lubyova, and P. M. Pitha. 2002. On the role of IRF in host defense. J. Interferon Cytokine Res. 22:59-71. - PubMed
-
- Basler, C. F., A. H. Reid, J. K. Dybing, T. A. Janczewski, T. G. Fanning, H. Zheng, M. Salvatore, M. L. Perdue, D. E. Swayne, A. Garcia-Sastre, P. Palese, and J. K. Taubenberger. 2001. Sequence of the 1918 pandemic influenza virus nonstructural gene (NS) segment and characterization of recombinant viruses bearing the 1918 NS genes. Proc. Natl. Acad. Sci. USA 98:2746-2751. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

