The human kinetochore proteins Nnf1R and Mcm21R are required for accurate chromosome segregation
- PMID: 16932742
- PMCID: PMC1560365
- DOI: 10.1038/sj.emboj.7601293
The human kinetochore proteins Nnf1R and Mcm21R are required for accurate chromosome segregation
Erratum in
- EMBO J. 2009 May 6;28(9):1374
Abstract
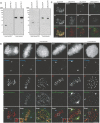
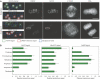
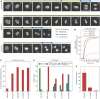
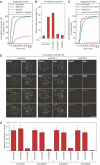
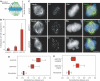
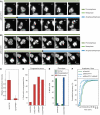
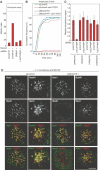
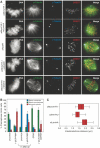
Kinetochores (KTs) assemble on centromeric DNA, bi-orient paired sister chromatids on spindle microtubules (MTs) and control cell-cycle progression via the spindle assembly checkpoint. Genetic and biochemical studies in budding yeast have established that three 'linker' complexes, MIND, COMA and NDC80, play essential but distinct roles in KT assembly and chromosome segregation. To determine whether similar linker activities are present at human KTs, we have compared the functions of Nnf1R and Mcm21R, recently identified MIND and COMA subunits, and Nuf2R, a well-characterized NDC80 subunit. We find that the three proteins bind to KTs independent of each other and with distinct cell-cycle profiles. MT-KT attachment is aberrant in Nnf1R- and Mcm21R-depleted cells, whereas it is lost in the absence of Nuf2R. Defective attachments in Nnf1R-depleted cells prevent chromosome congression, whereas those in Mcm21R-depleted cells interfere with spindle assembly. All three human KT proteins are necessary for correct binding of spindle checkpoint proteins to KTs. The differing functions and KT-binding properties of Nnf1R, Mcm21R and Nuf2R suggest that, like their yeast counterparts, the proteins act independent of each other in KT assembly, but that their combined activities are required for checkpoint signaling.
Figures



References
-
- Buffin E, Lefebvre C, Huang J, Gagou ME, Karess RE (2005) Recruitment of Mad2 to the kinetochore requires the Rod/Zw10 complex. Curr Biol 15: 856–861 - PubMed
-
- Chan GK, Liu ST, Yen TJ (2005) Kinetochore structure and function. Trends Cell Biol 15: 589–598 - PubMed
-
- Ciferri C, De Luca J, Monzani S, Ferrari KJ, Ristic D, Wyman C, Stark H, Kilmartin J, Salmon ED, Musacchio A (2005) Architecture of the human ndc80-hec1 complex, a critical constituent of the outer kinetochore. J Biol Chem 280: 29088–29095 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

