Genetic structure of wild and cultivated olives in the central Mediterranean basin
- PMID: 16935868
- PMCID: PMC2803593
- DOI: 10.1093/aob/mcl178
Genetic structure of wild and cultivated olives in the central Mediterranean basin
Abstract
Background and aims: Olive cultivars and their wild relatives (oleasters) represent two botanical varieties of Olea europaea subsp. europaea (respectively europaea and sylvestris). Olive cultivars have undergone human selection and their area of diffusion overlaps that of oleasters. Populations of genuine wild olives seem restricted to isolated areas of Mediterranean forests, while most other wild-looking forms of olive may include feral forms that escaped cultivation.
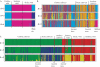
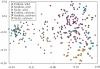
Methods: The genetic structure of wild and cultivated olive tree populations was evaluated by amplified fragment length polymorphism (AFLP) markers at a microscale level in one continental and two insular Italian regions.
Key results: The observed patterns of genetic variation were able to distinguish wild from cultivated populations and continental from insular regions. Island oleasters were highly similar to each other and were clearly distinguishable from those of continental regions. Ancient cultivated material from one island clustered with the wild plants, while the old plants from the continental region clustered with the cultivated group.
Conclusions: On the basis of these results, we can assume that olive trees have undergone a different selection/domestication process in the insular and mainland regions. The degree of differentiation between oleasters and cultivated trees on the islands suggests that all cultivars have been introduced into these regions from the outside, while the Umbrian cultivars have originated either by selection from local oleasters or by direct introduction from other regions.
Figures

Similar articles
-
Allozyme variation of oleaster populations (wild olive tree) (Olea europaea L.) in the Mediterranean Basin.Heredity (Edinb). 2004 Apr;92(4):343-51. doi: 10.1038/sj.hdy.6800430. Heredity (Edinb). 2004. PMID: 14985782
-
Genetic diversity and population structure of wild olives from the North-Western Mediterranean assessed by SSR markers.Ann Bot. 2007 Sep;100(3):449-58. doi: 10.1093/aob/mcm132. Epub 2007 Jul 5. Ann Bot. 2007. PMID: 17613587 Free PMC article.
-
Mitochondrial DNA variation and RAPD mark oleasters, olive and feral olive from Western and Eastern Mediterranean.Theor Appl Genet. 2002 May;104(6-7):1209-1216. doi: 10.1007/s00122-002-0883-7. Epub 2002 Apr 5. Theor Appl Genet. 2002. PMID: 12582632
-
The history of olive cultivation in the southern Levant.Front Plant Sci. 2023 Feb 23;14:1131557. doi: 10.3389/fpls.2023.1131557. eCollection 2023. Front Plant Sci. 2023. PMID: 36909452 Free PMC article. Review.
-
Plant-parasitic nematodes associated with olive tree (Olea europaea L.) with a focus on the Mediterranean Basin: a review.C R Biol. 2014 Jul-Aug;337(7-8):423-42. doi: 10.1016/j.crvi.2014.05.006. Epub 2014 Jul 26. C R Biol. 2014. PMID: 25103828 Review.
Cited by
-
Association Study of the 5'UTR Intron of the FAD2-2 Gene With Oleic and Linoleic Acid Content in Olea europaea L.Front Plant Sci. 2020 Feb 13;11:66. doi: 10.3389/fpls.2020.00066. eCollection 2020. Front Plant Sci. 2020. PMID: 32117401 Free PMC article.
-
High genetic diversity detected in olives beyond the boundaries of the Mediterranean Sea.PLoS One. 2014 Apr 7;9(4):e93146. doi: 10.1371/journal.pone.0093146. eCollection 2014. PLoS One. 2014. PMID: 24709858 Free PMC article.
-
Molecular and phytochemical analysis of wild type and olive cultivars grown under Saudi Arabian environment.3 Biotech. 2017 Oct;7(5):289. doi: 10.1007/s13205-017-0920-5. Epub 2017 Aug 24. 3 Biotech. 2017. PMID: 28868216 Free PMC article.
-
Wild and cultivated olive tree genetic diversity in Greece: a diverse resource in danger of erosion.Front Genet. 2023 Dec 4;14:1298565. doi: 10.3389/fgene.2023.1298565. eCollection 2023. Front Genet. 2023. PMID: 38111682 Free PMC article.
-
Genetic diversity and gene flow in a Caribbean tree Pterocarpus officinalis Jacq.: a study based on chloroplast and nuclear microsatellites.Genetica. 2009 Mar;135(2):185-98. doi: 10.1007/s10709-008-9268-4. Epub 2008 Apr 23. Genetica. 2009. PMID: 18431679
References
-
- Alcantara JM and Rey PJ. (2003) Conflicting selection pressures on seed size: evolutionary ecology of fruit size in a bird-dispersed tree. Olea europaea. Journal of Evolutionary Biology 161168–1176. - PubMed
-
- Angiolillo A, Mencuccini M, Baldoni L. (1999) Olive genetic diversity assessed using amplified fragment length polymorphisms. Theoretical and Applied Genetics 98411–421.
-
- Baldoni L, Pellegrini M, Mencuccini M, Angiolillo A, Mulas M. (2000) Genetic relationships among cultivated and wild olives revealed by AFLP markers. Acta Horticulturae 521275–284.
-
- Banilas G, Minas J, Gregoriu C, Demoliou C, Kourti A, Hatzopoulos P. (2003) Genetic diversity among accessions of an ancient olive variety of Cyprus. Genome 46370–376. - PubMed
-
- Belaj A, Rallo L, Trujillo I, Baldoni L. (2004) Using RAPD and AFLP markers to distinguish individuals obtained by clonal selection of ‘Arbequina’ and ‘Manzanilla de Sevilla’ olive. Hortscience 391566–1570.

