Regulation of bacterial priming and daughter strand synthesis through helicase-primase interactions
- PMID: 16935873
- PMCID: PMC1616961
- DOI: 10.1093/nar/gkl363
Regulation of bacterial priming and daughter strand synthesis through helicase-primase interactions
Abstract
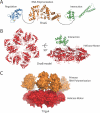
The replisome is a multi-component molecular machine responsible for rapidly and accurately copying the genome of an organism. A central member of the bacterial replisome is DnaB, the replicative helicase, which separates the parental duplex to provide templates for newly synthesized daughter strands. A unique RNA polymerase, the DnaG primase, associates with DnaB to repeatedly initiate thousands of Okazaki fragments per replication cycle on the lagging strand. A number of studies have shown that the stability and frequency of the interaction between DnaG and DnaB determines Okazaki fragment length. More recent work indicates that each DnaB hexamer associates with multiple DnaG molecules and that these primases can coordinate with one another to regulate their activities at a replication fork. Together, disparate lines of evidence are beginning to suggest that Okazaki fragment initiation may be controlled in part by crosstalk between multiple primases bound to the helicase.
Figures

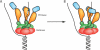
Similar articles
-
Mechanism and stoichiometry of interaction of DnaG primase with DnaB helicase of Escherichia coli in RNA primer synthesis.J Biol Chem. 2003 Dec 26;278(52):52253-61. doi: 10.1074/jbc.M308956200. Epub 2003 Oct 13. J Biol Chem. 2003. PMID: 14557266
-
Homogenous assays for Escherichia coli DnaB-stimulated DnaG primase and DnaB helicase and their use in screening for chemical inhibitors.Anal Biochem. 2002 May 15;304(2):174-9. doi: 10.1006/abio.2002.5627. Anal Biochem. 2002. PMID: 12009693
-
Identification of a region of Escherichia coli DnaB required for functional interaction with DnaG at the replication fork.J Biol Chem. 2000 Aug 25;275(34):26187-95. doi: 10.1074/jbc.M001800200. J Biol Chem. 2000. PMID: 10833513
-
The E. coli DNA Replication Fork.Enzymes. 2016;39:31-88. doi: 10.1016/bs.enz.2016.04.001. Epub 2016 May 13. Enzymes. 2016. PMID: 27241927 Review.
-
Replication Initiation in Bacteria.Enzymes. 2016;39:1-30. doi: 10.1016/bs.enz.2016.03.001. Epub 2016 Apr 20. Enzymes. 2016. PMID: 27241926 Free PMC article. Review.
Cited by
-
The conformational changes coupling ATP hydrolysis and translocation in a bacterial DnaB helicase.Nat Commun. 2019 Jan 3;10(1):31. doi: 10.1038/s41467-018-07968-3. Nat Commun. 2019. PMID: 30604765 Free PMC article.
-
The application of thermophilic DNA primase TtDnaG2 to DNA amplification.Sci Rep. 2017 Oct 9;7(1):12809. doi: 10.1038/s41598-017-12241-6. Sci Rep. 2017. PMID: 28993626 Free PMC article.
-
During heat stress in Myxococcus xanthus, the CdbS PilZ domain protein, in concert with two PilZ-DnaK chaperones, perturbs chromosome organization and accelerates cell death.PLoS Genet. 2023 Jun 20;19(6):e1010819. doi: 10.1371/journal.pgen.1010819. eCollection 2023 Jun. PLoS Genet. 2023. PMID: 37339150 Free PMC article.
-
Lambda gpP-DnaB Helicase Sequestration and gpP-RpoB Associated Effects: On Screens for Auxotrophs, Selection for Rif(R), Toxicity, Mutagenicity, Plasmid Curing.Viruses. 2016 Jun 22;8(6):172. doi: 10.3390/v8060172. Viruses. 2016. PMID: 27338450 Free PMC article.
-
Nutritional control of elongation of DNA replication by (p)ppGpp.Cell. 2007 Mar 9;128(5):865-75. doi: 10.1016/j.cell.2006.12.043. Cell. 2007. PMID: 17350574 Free PMC article.
References
-
- Kornberg A., Baker T. DNA Replication. 2nd edn. New York: Freeman; 1992.
-
- Rowen L., Kornberg A. Primase, the dnaG protein of Escherichia coli. An enzyme which starts DNA chains. J. Biol. Chem. 1978;253:758–764. - PubMed
-
- Bouche J.P., Zechel K., Kornberg A. dnaG gene product, a rifampicin-resistant RNA polymerase, initiates the conversion of a single-stranded coliphage DNA to its duplex replicative form. J. Biol. Chem. 1975;250:5995–6001. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

