Regulation of bacterial priming and daughter strand synthesis through helicase-primase interactions
- PMID: 16935873
- PMCID: PMC1616961
- DOI: 10.1093/nar/gkl363
Regulation of bacterial priming and daughter strand synthesis through helicase-primase interactions
Abstract
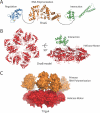
The replisome is a multi-component molecular machine responsible for rapidly and accurately copying the genome of an organism. A central member of the bacterial replisome is DnaB, the replicative helicase, which separates the parental duplex to provide templates for newly synthesized daughter strands. A unique RNA polymerase, the DnaG primase, associates with DnaB to repeatedly initiate thousands of Okazaki fragments per replication cycle on the lagging strand. A number of studies have shown that the stability and frequency of the interaction between DnaG and DnaB determines Okazaki fragment length. More recent work indicates that each DnaB hexamer associates with multiple DnaG molecules and that these primases can coordinate with one another to regulate their activities at a replication fork. Together, disparate lines of evidence are beginning to suggest that Okazaki fragment initiation may be controlled in part by crosstalk between multiple primases bound to the helicase.
Figures

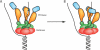
References
-
- Kornberg A., Baker T. DNA Replication. 2nd edn. New York: Freeman; 1992.
-
- Rowen L., Kornberg A. Primase, the dnaG protein of Escherichia coli. An enzyme which starts DNA chains. J. Biol. Chem. 1978;253:758–764. - PubMed
-
- Bouche J.P., Zechel K., Kornberg A. dnaG gene product, a rifampicin-resistant RNA polymerase, initiates the conversion of a single-stranded coliphage DNA to its duplex replicative form. J. Biol. Chem. 1975;250:5995–6001. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

