Role of RNA helicases in HIV-1 replication
- PMID: 16935887
- PMCID: PMC1616970
- DOI: 10.1093/nar/gkl398
Role of RNA helicases in HIV-1 replication
Abstract
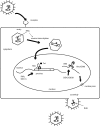
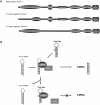
Viruses are replication competent genomes which are relatively gene-poor. Even the largest viruses (i.e. Herpesviruses) encode only slightly >200 open reading frames (ORFs). However, because viruses replicate obligatorily inside cells, and considering that evolution may be driven by a principle of economy of scale, it is reasonable to surmise that many viruses have evolved the ability to co-opt cell-encoded proteins to provide needed surrogate functions. An in silico survey of viral sequence databases reveals that most positive-strand and double-stranded RNA viruses have ORFs for RNA helicases. On the other hand, the genomes of retroviruses are devoid of virally-encoded helicase. Here, we review in brief the notion that the human immunodeficiency virus (HIV-1) has adopted the ability to use one or more cellular RNA helicases for its replicative life cycle.
Figures
Similar articles
-
A method to study the role of DDX3 RNA helicase in HIV-1.Methods Mol Biol. 2010;587:281-9. doi: 10.1007/978-1-60327-355-8_20. Methods Mol Biol. 2010. PMID: 20225157 Free PMC article.
-
Alterations in the expression of DEAD-box and other RNA binding proteins during HIV-1 replication.Retrovirology. 2004 Dec 8;1:42. doi: 10.1186/1742-4690-1-42. Retrovirology. 2004. PMID: 15588285 Free PMC article.
-
Cellular RNA helicases and HIV-1: insights from genome-wide, proteomic, and molecular studies.Virus Res. 2013 Feb;171(2):357-65. doi: 10.1016/j.virusres.2012.06.022. Epub 2012 Jul 16. Virus Res. 2013. PMID: 22814432 Free PMC article. Review.
-
From promoting to inhibiting: diverse roles of helicases in HIV-1 Replication.Retrovirology. 2012 Sep 28;9:79. doi: 10.1186/1742-4690-9-79. Retrovirology. 2012. PMID: 23020886 Free PMC article. Review.
-
Human Enterovirus Nonstructural Protein 2CATPase Functions as Both an RNA Helicase and ATP-Independent RNA Chaperone.PLoS Pathog. 2015 Jul 28;11(7):e1005067. doi: 10.1371/journal.ppat.1005067. eCollection 2015 Jul. PLoS Pathog. 2015. PMID: 26218680 Free PMC article.
Cited by
-
Characterization of staufen1 ribonucleoproteins by mass spectrometry and biochemical analyses reveal the presence of diverse host proteins associated with human immunodeficiency virus type 1.Front Microbiol. 2012 Oct 25;3:367. doi: 10.3389/fmicb.2012.00367. eCollection 2012. Front Microbiol. 2012. PMID: 23125841 Free PMC article.
-
DDX17 Specifically, and Independently of DDX5, Controls Use of the HIV A4/5 Splice Acceptor Cluster and Is Essential for Efficient Replication of HIV.J Mol Biol. 2018 Sep 14;430(18 Pt B):3111-3128. doi: 10.1016/j.jmb.2018.06.052. Epub 2018 Jul 2. J Mol Biol. 2018. PMID: 30131116 Free PMC article.
-
Ring expanded nucleoside analogues inhibit RNA helicase and intracellular human immunodeficiency virus type 1 replication.J Med Chem. 2008 Aug 28;51(16):5043-51. doi: 10.1021/jm800332m. Epub 2008 Aug 5. J Med Chem. 2008. PMID: 18680273 Free PMC article.
-
Knockdown of cellular RNA helicase DDX3 by short hairpin RNAs suppresses HIV-1 viral replication without inducing apoptosis.Mol Biotechnol. 2008 Jul;39(3):231-8. doi: 10.1007/s12033-008-9040-0. Epub 2008 Feb 8. Mol Biotechnol. 2008. PMID: 18259889
-
Template role of double-stranded RNA in tombusvirus replication.J Virol. 2014 May;88(10):5638-51. doi: 10.1128/JVI.03842-13. Epub 2014 Mar 5. J Virol. 2014. PMID: 24600009 Free PMC article.
References
-
- Caruthers J.M., McKay D.B. Helicase structure and mechanism. Curr. Opin. Struct. Biol. 2002;12:123–133. - PubMed
-
- Linder P., Lasko P.F., Ashburner M., Leroy P., Nielsen P.J., Nishi K., Schnier J., Slonimski P.P. Birth of the D-E-A-D box. Nature. 1989;337:121–122. - PubMed
-
- Rocak S., Linder P. DEAD-box proteins: the driving forces behind RNA metabolism. Nature Rev. Mol. Cell Biol. 2004;5:232–241. - PubMed
-
- Lorsch J.R. RNA chaperones exist and DEAD box proteins get a life. Cell. 2002;109:797–800. - PubMed

