Controlled expression and functional analysis of iron-sulfur cluster biosynthetic components within Azotobacter vinelandii
- PMID: 16936042
- PMCID: PMC1636278
- DOI: 10.1128/JB.00596-06
Controlled expression and functional analysis of iron-sulfur cluster biosynthetic components within Azotobacter vinelandii
Abstract
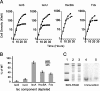
A system for the controlled expression of genes in Azotobacter vinelandii by using genomic fusions to the sucrose catabolic regulon was developed. This system was used for the functional analysis of the A. vinelandii isc genes, whose products are involved in the maturation of [Fe-S] proteins. For this analysis, the scrX gene, contained within the sucrose catabolic regulon, was replaced by the contiguous A. vinelandii iscS, iscU, iscA, hscB, hscA, fdx, and iscX genes, resulting in duplicate genomic copies of these genes: one whose expression is directed by the normal isc regulatory elements (Pisc) and the other whose expression is directed by the scrX promoter (PscrX). Functional analysis of [Fe-S] protein maturation components was achieved by placing a mutation within a particular Pisc-controlled gene with subsequent repression of the corresponding PscrX-controlled component by growth on glucose as the carbon source. This experimental strategy was used to show that IscS, IscU, HscBA, and Fdx are essential in A. vinelandii and that their depletion results in a deficiency in the maturation of aconitase, an enzyme that requires a [4Fe-4S] cluster for its catalytic activity. Depletion of IscA results in a null growth phenotype only when cells are cultured under conditions of elevated oxygen, marking the first null phenotype associated with the loss of a bacterial IscA-type protein. Furthermore, the null growth phenotype of cells depleted of HscBA could be partially reversed by culturing cells under conditions of low oxygen. Conserved amino acid residues within IscS, IscU, and IscA that are essential for their respective functions and/or whose replacement results in a partial or complete dominant-negative growth phenotype were also identified using this system.
Figures




References
-
- Agar, J. N., P. Yuvaniyama, R. F. Jack, V. L. Cash, A. D. Smith, D. R. Dean, and M. K. Johnson. 2000. Modular organization and identification of a mononuclear iron-binding site within the NifU protein. J. Biol. Inorg. Chem. 5:167-177. - PubMed
-
- Agar, J. N., L. Zheng, V. L. Cash, D. R. Dean, and M. K. Johnson. 2000. Role of the IscU protein in iron-sulfur cluster biosynthesis: IscS-mediated assembly of a [2Fe-2S] cluster in IscU. J. Am. Chem. Soc. 122:2136-2137.
-
- Ali, V., Y. Shigeta, U. Tokumoto, Y. Takahashi, and T. Nozaki. 2004. An intestinal parasitic protist, Entamoeba histolytica, possesses a non-redundant nitrogen fixation-like system for iron-sulfur cluster assembly under anaerobic conditions. J. Biol. Chem. 279:16863-16874. - PubMed
-
- Barras, F., L. Loiseau, and B. Py. 2005. How Escherichia coli and Saccharomyces cerevisiae build Fe/S proteins. Adv. Microb. Physiol. 50:41-101. - PubMed
-
- Beinert, H., R. H. Holm, and E. Munck. 1997. Iron-sulfur clusters: nature's modular, multipurpose structures. Science 277:653-659. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

