Apoptosis resistance downstream of eIF4E: posttranscriptional activation of an anti-apoptotic transcript carrying a consensus hairpin structure
- PMID: 16936314
- PMCID: PMC1636353
- DOI: 10.1093/nar/gkl558
Apoptosis resistance downstream of eIF4E: posttranscriptional activation of an anti-apoptotic transcript carrying a consensus hairpin structure
Abstract
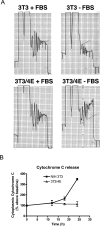
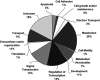
Aberrant activation of the translation initiation machinery is a common property of malignant cells, and is essential for breast carcinoma cells to manifest a malignant phenotype. How does sustained activation of the rate limiting step in protein synthesis so fundamentally alter a cell? In this report, we test the post transcriptional operon theory as a possible mechanism, employing a model system in which apoptosis resistance is conferred on NIH 3T3 cells by ectopic expression of eIF4E. We show (i) there is a set of 255 transcripts that manifest an increase in translational efficiency during eIF4E-mediated escape from apoptosis; (ii) there is a novel prototype 55 nt RNA consensus hairpin structure that is overrepresented in the 5'-untranslated region of translationally activated transcripts; (iii) the identified consensus hairpin structure is sufficient to target a reporter mRNA for translational activation under pro-apoptotic stress, but only when eIF4E is deregulated; and (iv) that osteopontin, one of the translationally activated transcripts harboring the identified consensus hairpin structure functions as one mediator of the apoptosis resistance seen in our model. Our findings offer genome-wide insights into the mechanism of eIF4E-mediated apoptosis resistance and provide a paradigm for the systematic study of posttranscriptional control in normal biology and disease.
Figures

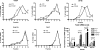

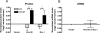
References
-
- Keene J.D., Lager P.J. Post-transcriptional operons and regulons co-ordinating gene expression. Chromosome Res. 2005;13:327–337. - PubMed
-
- Dever T.E. Gene-specific regulation by general translation factors. Cell. 2002;108:545–556. - PubMed
-
- Preiss T., Hentze M.W. Starting the protein synthesis machine: eukaryotic translation initiation. Bioessays. 2003;25:1201–1211. - PubMed
-
- Hershey J., Merrick W. The pathway and mechanism of protein synthesis. In: Sonenberg N., Hershey J., Mathews M., editors. Translational Control of Gene Expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 33–88.
-
- Clark I.E., Wyckoff D., Gavis E.R. Synthesis of the posterior determinant Nanos is spatially restricted by a novel cotranslational regulatory mechanism. Curr. Biol. 2000;10:1311–1314. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

