The retinoblastoma tumor-suppressor gene, the exception that proves the rule
- PMID: 16936742
- PMCID: PMC2799241
- DOI: 10.1038/sj.onc.1209616
The retinoblastoma tumor-suppressor gene, the exception that proves the rule
Abstract
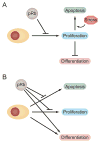
The retinoblastoma tumor-suppressor gene (Rb1) is centrally important in cancer research. Mutational inactivation of Rb1 causes the pediatric cancer retinoblastoma, while deregulation of the pathway in which it functions is common in most types of human cancer. The Rb1-encoded protein (pRb) is well known as a general cell cycle regulator, and this activity is critical for pRb-mediated tumor suppression. The main focus of this review, however, is on more recent evidence demonstrating the existence of additional, cell type-specific pRb functions in cellular differentiation and survival. These additional functions are relevant to carcinogenesis suggesting that the net effect of Rb1 loss on the behavior of resulting tumors is highly dependent on biological context. The molecular mechanisms underlying pRb functions are based on the cellular proteins it interacts with and the functional consequences of those interactions. Better insight into pRb-mediated tumor suppression and clinical exploitation of pRb as a therapeutic target will require a global view of the complex, interdependent network of pocket protein complexes that function simultaneously within given tissues.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

