Estrogen receptor (ER)-beta isoforms: a key to understanding ER-beta signaling
- PMID: 16938840
- PMCID: PMC1552044
- DOI: 10.1073/pnas.0605676103
Estrogen receptor (ER)-beta isoforms: a key to understanding ER-beta signaling
Erratum in
- Proc Natl Acad Sci U S A. 2006 Oct 3;103(40):14977
Abstract
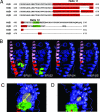
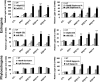
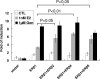
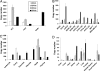
Estrogen receptor beta (ER-beta) regulates diverse physiological functions in the human body. Current studies are confined to ER-beta1, and the functional roles of isoforms 2, 4, and 5 remain unclear. Full-length ER-beta4 and -beta5 isoforms were obtained from a prostate cell line, and they exhibit differential expression in a wide variety of human tissues/cell lines. Through molecular modeling, we established that only ER-beta1 has a full-length helix 11 and a helix 12 that assumes an agonist-directed position. In ER-beta2, the shortened C terminus results in a disoriented helix 12 and marked shrinkage in the coactivator binding cleft. ER-beta4 and -beta5 completely lack helix 12. We further demonstrated that ER-beta1 is the only fully functional isoform, whereas ER-beta2, -beta4, and -beta5 do not form homodimers and have no innate activities of their own. However, the isoforms can heterodimerize with ER-beta1 and enhance its transactivation in a ligand-dependent manner. ER-beta1 tends to form heterodimers with other isoforms under the stimulation of estrogens but not phytoestrogens. Collectively, these data support the premise that (i) ER-beta1 is the obligatory partner of an ER-beta dimer, whereas the other isoforms function as variable dimer partners with enhancer activity, and (ii) a single functional helix 12 in a dimer is sufficient for gene transactivation. Thus, ER-beta behaves like a noncanonical type-I receptor, and its action may depend on differential amounts of ER-beta1 homo- and heterodimers formed upon stimulation by a specific ligand. Our findings have provided previously unrecognized directions for studying ER-beta signaling and design of ER-beta-based therapies.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

