Phylogenetic evidence for inter-typic recombination in the emergence of human enterovirus 71 subgenotypes
- PMID: 16939656
- PMCID: PMC1569848
- DOI: 10.1186/1471-2180-6-74
Phylogenetic evidence for inter-typic recombination in the emergence of human enterovirus 71 subgenotypes
Abstract
Background: Human enterovirus 71 (EV-71) is a common causative agent of hand, foot and mouth disease (HFMD). In recent years, the virus has caused several outbreaks with high numbers of deaths and severe neurological complications. Several new EV-71 subgenotypes were identified from these outbreaks. The mechanisms that contributed to the emergence of these subgenotypes are unknown.
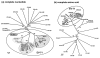
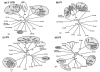
Results: Six EV-71 isolates from an outbreak in Malaysia, in 1997, were sequenced completely. These isolates were identified as EV-71 subgenotypes, B3, B4 and C2. A phylogenetic tree that correlated well with the present enterovirus classification scheme was established using these full genome sequences and all other available full genome sequences of EV-71 and human enterovirus A (HEV-A). Using the 5' UTR, P2 and P3 genomic regions, however, isolates of EV-71 subgenotypes B3 and C4 segregated away from other EV-71 subgenotypes into a cluster together with coxsackievirus A16 (CV-A16/G10) and EV-71 subgenotype C2 clustered with CV-A8. Results from the similarity plot analyses supported the clustering of these isolates with other HEV-A. In contrast, at the same genomic regions, a CV-A16 isolate, Tainan5079, clustered with EV-71. This suggests that amongst EV-71 and CV-A16, only the structural genes were conserved. The 3' end of the virus genome varied and consisted of sequences highly similar to various HEV-A viruses. Numerous recombination crossover breakpoints were identified within the non-structural genes of some of these newer EV-71 subgenotypes.
Conclusion: Phylogenetic evidence obtained from analyses of the full genome sequence supports the possible occurrence of inter-typic recombination involving EV-71 and various HEV-A, including CV-A16, the most common causal agent of HFMD. It is suggested that these recombination events played important roles in the emergence of the various EV-71 subgenotypes.
Figures


References
-
- Poyry T, Kinnunen L, Hyypia T, Brown B, Horsnell C, Hovi T, Stanway G. Genetic and phylogenetic clustering of enteroviruses. J Gen Virol. 1996;77:1699–1717. - PubMed
-
- Melnick JL. Enteroviruses: polioviruses, coxsackieviruses, echoviruses and newer enteroviruses. In: Fields BN, Knipe DM, Howley PM, Chanock RM, Melnick JL, Moath TP, Roizman B, Straus SE, editor. Fields Virology. 3. Philadelphia: Lippincott-Raven Publishers; 1996. pp. 655–711.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

